The management of infections secondary to multidrug-resistant organisms (MDROs) that encompasses gram-negative bacteria (GNB) is a global healthcare challenge not only because of the limited available treatment options but also for their associated significant morbidity and mortality as well as the substantial cost of management.1,2
In secondary and tertiary hospitals, the ultimate antimicrobial resistance (AMR) is encountered at intensive care units (ICUs) where the critical nature of patients’ cohort, concurrent comorbidities, invasive procedures, prior colonization as well as environmental exposure to MDROs that is accelerated by high antibiotics consumption are inevitable acquisition hazards.3,4 Over the past decade in Qatar, internal microbiological surveillance and monitoring of GNB, particularly the Enterobacterales, has revealed alarmingly rising trends of AMR, particularly for extended-spectrum β-lactamases (ESBLs) in line with shifting regional epidemiology.5,6 In critical care settings typical recommended approach for the management of ESBL-producing Enterobacterales is treatment with carbapenems, particularly if there is an associated serious invasive or high inoculum disease.7 The concern of diminishing efficacy of the limited treatment options against the ever-rising resistant bacterial strains, led infection specialists to seek alternatives to carbapenem therapy.
Ceftazidime/avibactam and ceftolozane/tazobactam are β-lactam/β-lactamase inhibitors (BLBLIs) combinations that are approved by both the United States Food and Drug Administration and the European Medicines Agency, demonstrating comparable or superior activity against MDROs particularly in GNB for the treatment of complicated urinary tract and intra-abdominal infections as well as infections secondary to hospital or ventilation associated pneumonia.8 Avibactam is a non-BLBLI that potently inhibits most (but not all) class A ESBLs, class C (including AmpC enzymes), and some class D β-lactamases.9 Furthermore, due to its different mode of action, avibactam is considered as one of the most effective BLBLIs displaying a broader inhibitory range and spectrum.10 On the other hand, ceftolozane is a novel cephalosporin that is not affected by outer membrane protein loss which is a weak substrate for drug efflux pump mechanism, rendering the drug exhibiting less affinity for hydrolysis by AmpC, and hence better efficacy.11 Pairing ceftolozane with the classic β-lactamase inhibitor tazobactam has broadened their capacity to act on most ESBL-producing GNB.12
The presented study aims mainly to evaluate the antimicrobial activity of ceftazidime/avibactam and ceftolozane/tazobactam against 109 ESBL-producing Enterobacterales isolates from ICUs in Qatar,13 describe its microbiological characteristics, as well as the underlying genomic resistance profiles.
Methods
This research project was approved by the Institutional Review Board at Hamad Medical Corporation (HMC), which complies with international ethical standards and regulations (Protocol no. RC/75813/2013). The study was conducted on routine specimens processed by the Microbiology Division, Department of Laboratory Medicine and Pathology, HMC, Qatar. All samples were collected prospectively over one year (1 November 2012 to 31 October 2013) from patients admitted to all ICUs (medical 29%, surgical 29%, trauma 16%, pediatric 16%, and neonatal 10%) at HMC. These were then analyzed for the presence of resistant pathogens.
The study definitions recognized duplicates of the same species of bacteria as isolates from the same patient displaying identical antimicrobial susceptibility patterns when isolated within 30 days regardless of sample sites which were considered repetitive and excluded. Isolates with major differences in antimicrobial susceptibilities were counted as new even within the defined 30 days time frame. The single isolate that was resistant to ceftazidime/avibactam and ceftolozane/tazobactam underwent standard diagnostic work-up, and then was stored at -80 °C pending further genomic analysis.
Microbiological identification and antimicrobial susceptibility tests (AST) were performed using BD PhoenixTM automated system according to manufacturer recommendations. Samples that tested positive for ESBL by Phoenix, or showed a minimum inhibitory concentration (MIC) of > 8 μg/mL for 3rd generation cephalosporins or aztreonam, were subsequently confirmed by a double-disk potentiation test with ceftazidime, amoxicillin/clavulanic acid, ceftriaxone, and cefoxitin antibiotics, interpreted as recommended by Clinical Laboratory Standards Institute standards for ESBL identification.14 AST and MIC for ceftazidime/avibactam and ceftolozane/tazobactam were performed using MIC Test Strips (Liofilchem®, Diagnostics, Italy). Escherichia coli ATCC 25922, E. coli ATCC 35218, and Pseudomonas aeruginosa ATCC 27853 were used as controls. Susceptibility reporting was based on the Clinical Laboratory Standards Institute recommendations.14 Since there were no recommended intermediate susceptibility categories available for ceftazidime/avibactam against Enterobacterales, isolates were described as ‘susceptible’ if the MIC was ≤ 8 mg/L and ‘non-susceptible’ if the MIC was > 8 mg/L as outlined in the supplementary Table A1 in Appendix.14 To achieve consistency, intermediate and resistant categories were grouped as non-susceptible for all reported antimicrobial agents.
Bacterial DNA extraction and detection of ESBL resistance genes were performed through an in-house polymerase chain reaction (PCR) technique, using the boiling lysis methods.15 PCR reactions for the ESBL genes (TEM, SHV, and CTX-M-1) were conducted using previously described protocols.16
Whole-genome sequencing (WGS) was performed to study isolated genomic relationships for annotating antibiotic resistance genes (ARGs). Extracted DNA was sent to GATC Service (Eurofins Genomics, Germany) for sequencing using Illumina HiSeq 2000 system (Illumina, San Diego, California). The genes were assembled using SPAdes, Version 3.13.0 (https://cab.spbu.ru/software/spades/) while Multi-locus sequence typing (MLST) of the described resistant isolate of E. coli was performed on MLST server 1.8 provided (https://cge.cbs.dtu.dk/services/MLST/). ARGs were annotated using Comprehensive Antibiotic Resistance Database (CARD), Version 1.2.0 (https://card.mcmaster.ca/).
Demographics of patients, characteristics of isolates, as well as the patterns of antimicrobial susceptibility of ESBL-producing Enterobacterales including resistant genes were presented as numbers and percentages using Stata statistical software (Stata Corp LLC, College Station, Texas version 16.1).
Results
Out of 629 Enterobacterales isolates investigated, 109 (17.3%) isolates from 87 patients were found to be ESBL positive. The source samples of these were: respiratory 35.8% (39/109), blood 27.5% (30/109), urine 24.8% (27/109), fluids 6.4% (7/109), and others 5.5% (6/109). The ESBL-positive isolates were predominantly Klebsiella pneumoniae (50.5%) and E. coli (34.9%) while other species comprised 13.7%. The majority of isolates were from male patients 65 (59.6%) and those aged between one month and 86 years. The patients were categorized into three age groups labeled adult, pediatric, and geriatrics. ‘Adult’ group (14–65 years) contributed to more than half (57/109; 52.3%) of the resistant isolates, followed by ‘pediatric’ (9/109; 8.3%) < 14 years, and ‘geriatric’ (26/109; 23.9%) > 65 years.
Table 1: Minimum inhibitory concentration (MIC) for ceftazidime/avibactam and ceftolozane/tazobactam against 109 clinical extended-spectrum β-lactamase-producing Enterobacterales isolates collected from intensive care units, Hamad Medical Corporation, Qatar
|
Klebsiella pneumoniae ssp pneumoniae
|
55
|
CZA
|
0.09–0.75
|
55 (100)
|
0.25
|
0.38
|
|
C/T
|
0.25–1.50
|
55 (100)
|
0.38
|
1.00
|
|
Escherichia coli
|
38
|
CZA
|
0.06–256.00
|
37 (97.4)
|
0.12
|
0.38
|
|
C/T
|
0.19–256.00
|
37 (97.4)
|
0.38
|
0.75
|
|
Enterobacter aerogenes
|
4
|
CZA
|
0.09–0.25
|
4 (100)
|
0.19
|
0.25
|
|
C/T
|
0.38–0.50
|
4 (100)
|
0.38
|
0.50
|
|
Enterobacter cloacae
|
4
|
CZA
|
0.09–0.19
|
4 (100)
|
0.09
|
0.19
|
|
C/T
|
0.19–0.25
|
4 (100)
|
0.25
|
0.25
|
|
Serratia marcescens
|
2
|
CZA
|
0.02–0.12
|
2 (100)
|
0.02
|
0.12
|
|
C/T
|
0.19
|
2 (100)
|
0.19
|
0.19
|
|
Citrobacter braakii
|
1
|
CZA
|
0.25
|
1 (100)
|
0.25
|
0.25
|
|
C/T
|
0.75
|
1 (100)
|
0.75
|
0.75
|
|
Citrobacter freundii
|
1
|
CZA
|
0.06
|
1 (100)
|
0.06
|
0.06
|
|
C/T
|
0.38
|
1 (100)
|
0.38
|
0.38
|
|
Citrobacter amalonaticus
|
1
|
CZA
|
0.12
|
1 (100)
|
0.12
|
0.12
|
|
C/T
|
0.19
|
1 (100)
|
0.19
|
0.19
|
|
Klebsiella oxytoca
|
1
|
CZA
|
0.12
|
1 (100)
|
0.12
|
0.12
|
|
C/T
|
0.38
|
1 (100)
|
0.38
|
0.38
|
|
Klebsiella pneumoniae ssp ozaenae
|
1
|
CZA
|
0.09
|
1 (100)
|
0.09
|
0.09
|
|
C/T
|
0.25
|
1 (100)
|
0.25
|
0.25
|
|
Proteus penneri
|
1
|
CZA
|
0.04
|
1 (100)
|
0.04
|
0.04
|
|
C/T
|
1.00
|
1 (100)
|
1.00
|
1.00
|
|
109
|
CZA
|
0.02–256.00
|
108 (99.1)
|
0.19
|
0.38
|
CZA: ceftazidime-avibactam; C/T: ceftolozane-tazobactam.
Table 2: Comparison of minimum inhibitory concentration for ceftazidime/avibactam vs ceftolozane/tazobactam against 109 clinical extended-spectrum β-lactamase-producing Enterobacterales isolates from samples from intensive care unit patients at Hamad Medical Corporation, Qatar.
< 0.1
|
13 (11.9)
|
6 (5.5)
|
1 (0.9)
|
1 (0.9)
|
0.0
|
21 (19.3)
|
|
< 0.25
|
7 (6.4)
|
13 (11.9)
|
0.0
|
0.0
|
0.0
|
20 (18.3)
|
|
< 0.5
|
5 (4.6)
|
37 (33.9)
|
14 (12.8)
|
7 (6.4)
|
0.0
|
63 (57.8)
|
|
< 0.75
|
0.0
|
0.0
|
0.0
|
3 (2.8)
|
0.0
|
3 (2.8)
|
|
< 1
|
0.0
|
1 (0.9)
|
0.0
|
0.0
|
0.0
|
1 (0.9)
|
|
> 256
|
0.0
|
0.0
|
0.0
|
0.0
|
1 (0.9)
|
1 (0.9)
|
Table 3: Summary of studies comparing in vitro activity of ceftazidime/avibactam and ceftolozane/tazobactam against extended-spectrum β-lactamase-producing Enterobacterales from different geographical regions worldwide.
|
Alatoom et al,17 2017
|
Abu Dhabi, UAE
|
Etest
|
Resistant to ≥ 1 agent from ≥ 3 antimicrobial classes
|
2015–2016
|
31
|
NA
|
29 (93.5)
|
30 (96.8)
|
|
Sader et al,18 2020
|
70 medical centers, USA
|
Broth microdilution
|
ESBL-producing Enterobacterales from patients hospitalized with pneumonia
|
2017–2018
|
285
|
283 (99.3)
|
285 (100)
|
219 (76.8)
|
|
Viala et al,19 2019
|
Montpellier, France
|
Etest
|
3rd G cephalosporin resistant Enterobacteriaceae
|
2017
|
62
|
NA
|
60 (97)
|
34 (65)
|
|
Araj et al,20 2020
|
Beirut, Lebanon
|
MIC gradient Strip Test
|
MDR and ESBLs E. coli and K. pneumoniae
|
2017–2018
|
199
|
NA
|
NA
|
159 (79.9)
|
CZA: ceftazidime/avibactam; C/T: ceftolozane/tazobactam; MDR: multi-drug resistant; MEM: meropenem; NS: non-susceptible.
*All studies reported the isolates as susceptible if the MIC was ≤ 8 mg/L for ceftazidime/avibactam and ≤ 4 mg/L for ceftolozane/tazobactam.
Table 4: Genotypic profiles of different β-lactamase enzymes detected among extended-spectrum β-lactamase-producing E. coli isolated from samples from intensive care unit patients at Hamad Medical Corporation, Qatar.
|
CTX-M-15
|
Class A β-lactamase
|
100
|
|
VEB-5
|
Class A β-lactamase
|
100
|
|
VIM-2
|
Class B β-lactamase
|
100
|
|
E. coli ampC
|
Class C β-lactamase
|
97.9
|
|
E. coli ampC1
|
Class C β-lactamase
|
99.3
|
|
E. coli ampH
|
Class C β-lactamase
|
99.2
|
|
CMY-42
|
Class C β-lactamase
|
100
|
|
OXA-10
|
Class D β-lactamase
|
100
|
|
OXA-4
|
Class D β-lactamase
|
100
|
The predominantly identified ESBL-producing genes were blaCTX-M-1 (72/109; 66.1%) followed by blaSHV (58/109; 53.2 %) and blaTEM (44/109; 40.4%). All three β-lactamase genes (TEM, SHV, and CTX-M-1) were detected in 26/56 (46.4%) of K. pneumoniae isolates, while two genes (SHV/CTX-M-1) were present in 10/56 (17.8%) of
K. pneumoniae and only 1/38 (2.6%) of E. coli isolates, with TEM/CTX-M-1 being present in 7/38 (18.4%) of E. coli and 4/56 (7.1%) of K. pneumoniae, while TEM/SHV was detected in only 2/38 (5.3%) of E. coli isolates.
The activity of ceftazidime/avibactam and ceftolozane/tazobactam against 109 ESBL-producing Enterobacterales isolates demonstrated 99.1% (108/109) susceptibility for both combinations. Only meropenem showed 100% (109/109) susceptibility followed by imipenem at 99.1% while ertapenem and amikacin susceptibility was 97.2%. Other antimicrobials demonstrated moderate-to-low susceptibility rates with 78.0% for piperacillin/tazobactam, 64.2% for tigecycline, 60.6% for ciprofloxacin, and 38.5% for co-trimoxazole while as predicted cephalosporin had high-level resistance (99.1% for ceftriaxone and 93.6% for cefepime) [Figure 1]. Furthermore, most of the ESBL-producing Enterobacterales were highly susceptible to ceftazidime/avibactam at low MICs (MIC50/90 0.19/0.38 µg/mL) and ceftolozane/tazobactam (MIC50/90 0.38/1.00 µg/mL) [Table 1], with the majority of isolates demonstrating MICs < 0.5 (n = 81; 74.3%) [Table 2]. The additional microbiological and molecular characterization including susceptibility testing results are shown in Appendix [Table A1].

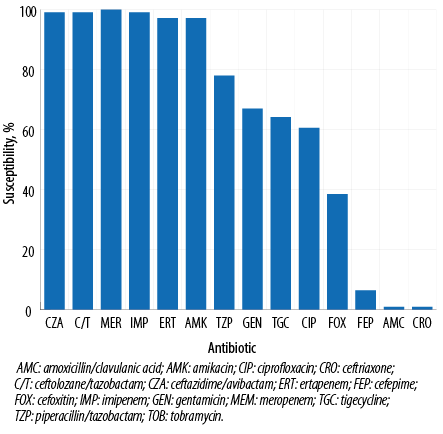 Figure 1: Antimicrobial susceptibility results for ceftazidime/avibactam, ceftolozane/tazobactam, and comparator agents against clinical extended-spectrum β-lactamase-producing Enterobacterales isolates from Qatar.
Figure 1: Antimicrobial susceptibility results for ceftazidime/avibactam, ceftolozane/tazobactam, and comparator agents against clinical extended-spectrum β-lactamase-producing Enterobacterales isolates from Qatar.
Our findings are distinctively different from other regional studies where ceftazidime/avibactam demonstrated superior activity when compared to ceftolozane/tazobactam against ESBL-producer [Table 3], which suggests a potential correlation of embedded ESBL resistance genes not demonstrated in our study because of paucity of resistant
isolates [Table 4].22
Among the 109 identified ESBL-producing Enterobacteralesw only one (0.9%) E. coli isolate was completely resistant to both ceftolozane/tazobactam and ceftolozane/tazobactam, with MIC > 256 [Table 1]. The resistant isolate was collected from peritoneal fluid of a fatal case of complicated intra-abdominal infection, and was subsequently identified as sequence type ST38. Genomic data analysis revealed that the resistant isolate possessed different ARGs including 11 different β-lactamase genes from all classes; Class A ESBL (CTX-M-1 and VEB-5), Class B metallo-β-lactamase (MBL) including blaVIM-2, class C β-lactamase including blaPCMY-42. Class D β-lactamase such as blaOXA-4, blaOXA-10, and blaOXA-486 [Table 4].
Discussion
AMR is a major global healthcare challenge with ominous outcomes. Its ultimate manifestation occurs at critical care units where potent risk factors converge—such as a hazardous environment, vulnerable host, and highly resistant pathogens.23 Thus, one of the foremost challenges in critical care is prevention and management of infections caused by MDR gram-negative organisms, particularly ESBL-producing Enterobacterales resistant to most antimicrobial classes including most β-lactam penicillins, BLBLIs, and cephalosporins.23,24 To combat the growing problem of ESBL-producing Enterobacterales, recent decades have witnessed exponentially rising reliance on carbapenems, to the point of their becoming the sine qua non for its management, especially in the context of invasive or high-burden disease.23,25 In complicated ESBL infections, randomized control trials have demonstrated the superiority of carbapenems over comparators including BLBIs.25 However, the near-universal use of carbapenems has not prevented the problem from increasing; hence, the relentless search for ever-more powerful antibiotic regimens.8 Among the most promising new combinations are ceftazidime/avibactam and ceftolozane/tazobactam. Several global studies have evaluated the spectrum of their efficacy in vitro and in vivo against highly resistant gram-negative strains including ESBL producers and their regional variations.17–21,26
A 2010 study in Qatar that investigated 450 episodes of invasive bacteremia from a single institution found 61% prevalence of GNB, the most prevalent of which were E. coli (27.8%) and K. pneumoniae (17.9%), most of them ESBL producers.27 The scale of the growing problem at the same institution became clearer in a 2020 study which analyzed the results of culture-positive complicated urinary tract infections among adults admitted to surgical ICUs over a 10-year period (2008–2018). The study found that 36% of the isolated pathogens were ESBLs.28 Healthcare leaders in the Gulf Cooperation Council countries have recognized the problem of resistant pathogens as a health priority and initiated regional collaboration against this common threat.29
In our study, 99.1% of ESBL isolates were highly susceptible and most isolates (74.3%) exhibited MIC < 0.5 for both ceftazidime/avibactam and ceftolozane/tazobactam [Table 2]. Notably, the observed high-level susceptibility for ceftazidime/avibactam and ceftolozane/tazobactam against ESBL-producing Enterobacterales isolates collected from prospective critical care clinical cases, predates the introduction of these agents into clinical practice in Qatar. Our microbiological evaluation suggests these novel agents might be rational empirical treatment options sparing carbapenems.
In the Arabian Gulf region, the high volume of international travel coupled with population diversity and high antibiotic consumption are contributing factors towards the rising and diversifying trends of ESBLs in GNB. The problem has reached an endemic state requiring alternative management options.6,30
In line with regional and global ESBL genomic studies, the observation that blaCTX-M in conjunction with blaSHV and blaTEM are the main ARGs for ESBL-producing Enterobacterales, points towards the role of cephalosporins as the potential driving precipitant.5,21,31 In Qatar, the molecular epidemiology of Enterobacterales from the pediatric population follows the same trends in the region when a large study of 327 sequenced ESBL producers from clinical samples at the largest children's hospital in the region demonstrated dominance of E. coli and K. pneumoniae as main pathogens with predominance of balCTX-M-1 and coproduction of blaOXA-1 and blaTEM-1B as ARGs.32 In contrast, in the adults population, there are no detailed recent studies to evaluate the wider molecular epidemiology of ESBL in the country but the study of 149 non-repetitive carbapenem-resistant Enterobacterales confirmed regional preponderance of blaNDM and blaOXA48.33
Not surprisingly, following undergoing WGS, the only concomitant isolate resistant to both ceftazidime/avibactam and ceftolozane/tazobactam harbored multitude of different ARGs.
The ESBL-producing E. coli which belonged to ST38 possessed β-lactamase genes from all classes as shown in Table 3. Intriguingly, the detailed study demonstrated the presence of blaVIM-2 MBL which is known to play a fundamental role in ceftazidime/avibactam and ceftolozane/tazobactam resistance.34 In addition to the endemic class A blaCTX-M the resistant isolate also harbored blaVEB-5, which was initially detected in E. coli in the USA (GenBank accession number EF420108). The ARG, blaVEB confers high-level resistance to cephalosporins as well as monobactams and has been shown to inactivate ceftolozane/tazobactam.35 However, blaVEB-5 is known to be inhibited by avibactam which restored the MIC of ceftazidime from 256 μg/mL to 2 μg/mL for ceftazidime/avibactam combination.36 In addition to that, the resistant isolate has multiple underlying ARGs including blaPCMY-42 (AmpC), which drives ceftolozane/tazobactam resistance37 as well as class D β-lactamases blaOXA-10, which has been recently reported to enhance ceftolozane/tazobactam and ceftazidime/avibactam resistance.38
Despite its wide mechanism of action against MDROs including class A, C, and D β-lactamases, both ceftazidime/avibactam and ceftolozane/tazobactam remain vulnerable when encountering embedded class B β-lactamases such as the potent carbapenemase blaVIM-2 MBL, as described in the isolate of the study.21,39 Although some molecular tests have been developed to screen for ceftazidime/avibactam and ceftolozane/tazobactam resistance, the current recommendations to interpret activity through the golden routes of ASTs hold true.40
As a consequence, from our study, the prime recommendation is to conduct an urgent clinical evaluation of the novel antibiotics as alternative therapeutic options for MDROs including ESBLs, particularly in critical care settings. This should be supported by data from surveillance and monitoring mechanisms to evaluate the prevalence and characteristics of AMR in the region.
Conclusion
ESBL-producing Enterobacterales represent a significant and growing threat to healthcare in Qatar and the Arabian Gulf region in general, particularly in critical care settings. MDROs such as K. pneumoniae and E. coli harboring multiple ARGs continue to predominate. Promising high in vitro antimicrobial susceptibility to ceftazidime/avibactam and ceftolozane/tazobactam against ESBLs-producing Enterobacterales have added to the arsenal of alternative management options to overcome the growing resistance problem.
Disclosure
The authors declared no conflicts of interest. The publication of this article was funded by the Qatar National Library. Funders were not involved in the conduct of the study, preparation of the manuscript or the decision to submit the manuscript
for publication.
references
- 1. Naylor NR, Atun R, Zhu N, Kulasabanathan K, Silva S, Chatterjee A, et al. Estimating the burden of antimicrobial resistance: a systematic literature review. Antimicrob Resist Infect Control 2018 Apr;7:58.
- 2. Wozniak TM, Barnsbee L, Lee XJ, Pacella RE. Using the best available data to estimate the cost of antimicrobial resistance: a systematic review. Antimicrob Resist Infect Control 2019 Feb;8:26.
- 3. Massart N, Camus C, Benezit F, Moriconi M, Fillatre P, Le Tulzo Y. Incidence and risk factors for acquired colonization and infection due to extended-spectrum beta-lactamase-producing gram-negative bacilli: a retrospective analysis in three ICUs with low multidrug resistance rate. Eur J Clin Microbiol Infect Dis 2020 May;39(5):889-895.
- 4. Teerawattanapong N, Kengkla K, Dilokthornsakul P, Saokaew S, Apisarnthanarak A, Chaiyakunapruk N. Prevention and control of multidrug-resistant gram-negative bacteria in adult intensive care units: a systematic review and network meta-analysis. Clin Infect Dis 2017 May;64(suppl_2):S51-S60.
- 5. Zowawi HM, Balkhy HH, Walsh TR, Paterson DL. β-Lactamase production in key gram-negative pathogen isolates from the Arabian Peninsula. Clin Microbiol Rev 2013 Jul;26(3):361-380.
- 6. Dandachi I, Chaddad A, Hanna J, Matta J, Daoud Z. Understanding the epidemiology of multi-drug resistant gram-negative bacilli in the Middle East using a one health approach. Front Microbiol 2019 Aug;10:1941.
- 7. Rupp ME, Fey PD. Extended spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae: considerations for diagnosis, prevention and drug treatment. Drugs 2003;63(4):353-365.
- 8. van Duin D, Bonomo RA. Ceftazidime/Avibactam and Ceftolozane/Tazobactam: Second-generation β-Lactam/β-Lactamase Inhibitor Combinations. Clin Infect Dis 2016 Jul;63(2):234-241.
- 9. Nichols WW, Newell P, Critchley IA, Riccobene T, Das S. Avibactam pharmacokinetic/pharmacodynamic targets. Antimicrob Agents Chemother 2018 May;62(6):62.
- 10. Tuon FF, Rocha JL, Formigoni-Pinto MR. Pharmacological aspects and spectrum of action of ceftazidime-avibactam: a systematic review. Infection 2018 Apr;46(2):165-181.
- 11. Shortridge D, Pfaller MA, Castanheira M, Flamm RK. Antimicrobial activity of ceftolozane-tazobactam tested against Enterobacteriaceae and Pseudomonas aeruginosa collected from patients with bloodstream infections isolated in United States hospitals (2013-2015) as part of the Program to Assess Ceftolozane-Tazobactam Susceptibility (PACTS) surveillance program. Diagn Microbiol Infect Dis 2018 Oct;92(2):158-163.
- 12. Sucher AJ, Chahine EB, Cogan P, Fete M. Ceftolozane/tazobactam: a new cephalosporin and β-lactamase inhibitor combination. Ann Pharmacother 2015 Sep;49(9):1046-1056.
- 13. Sid Ahmed MA, Bansal D, Acharya A, Elmi AA, Hamid JM, Sid Ahmed AM, et al. Antimicrobial susceptibility and molecular epidemiology of extended-spectrum beta-lactamase-producing Enterobacteriaceae from intensive care units at Hamad Medical Corporation, Qatar. Antimicrob Resist Infect Control 2016 Feb;5:4.
- 14. Clinical Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. Wayne, PA, USA; 2020.
- 15. Queipo-Ortuño MI, De Dios Colmenero J, Macias M, Bravo MJ, Morata P. Preparation of bacterial DNA template by boiling and effect of immunoglobulin G as an inhibitor in real-time PCR for serum samples from patients with brucellosis. Clin Vaccine Immunol 2008 Feb;15(2):293-296.
- 16. Shaheen BW, Nayak R, Foley SL, Kweon O, Deck J, Park M, et al. Molecular characterization of resistance to extended-spectrum cephalosporins in clinical Escherichia coli isolates from companion animals in the United States. Antimicrob Agents Chemother 2011 Dec;55(12):5666-5675.
- 17. Alatoom A, Elsayed H, Lawlor K, AbdelWareth L, El-Lababidi R, Cardona L, et al. Comparison of antimicrobial activity between ceftolozane-tazobactam and ceftazidime-avibactam against multidrug-resistant isolates of Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa. Int J Infect Dis 2017 Sep;62:39-43.
- 18. Sader HS, Flamm RK, Carvalhaes CG, Castanheira M. Comparison of ceftazidime-avibactam and ceftolozane-tazobactam in vitro activities when tested against gram-negative bacteria isolated from patients hospitalized with pneumonia in United States medical centers (2017-2018). Diagn Microbiol Infect Dis 2020 Mar;96(3):114833.
- 19. Viala B, Zaidi FZ, Bastide M, Dumont Y, Le Moing V, Jean-Pierre H, et al. Assessment of the in vitro activities of ceftolozane/tazobactam and ceftazidime/avibactam in a collection of beta-lactam-resistant enterobacteriaceae and pseudomonas aeruginosa clinical isolates at Montpellier University Hospital, France. Microb Drug Resist 2019 Nov;25(9):1325-1329.
- 20. Araj GF, Berjawi DM, Musharrafieh U, El Beayni NK. Activity of ceftolozane/tazobactam against commonly encountered antimicrobial resistant Gram-negative bacteria in Lebanon. J Infect Dev Ctries 2020 Jun;14(6):559-564.
- 21. Hirsch EB, Brigman HV, Zucchi PC, Chen A, Anderson JC, Eliopoulos GM, et al. Ceftolozane-tazobactam and ceftazidime-avibactam activity against β-lactam-resistant Pseudomonas aeruginosa and extended-spectrum β-lactamase-producing Enterobacterales clinical isolates from U.S. medical centres. J Glob Antimicrob Resist 2020 Sep;22:689-694.
- 22. Ortiz de la Rosa J-M, Nordmann P, Poirel L. ESBLs and resistance to ceftazidime/avibactam and ceftolozane/tazobactam combinations in Escherichia coli and Pseudomonas aeruginosa. J Antimicrob Chemother 2019 Jul;74(7):1934-1939.
- 23. Bassetti M, Poulakou G, Timsit J-F. Focus on antimicrobial use in the era of increasing antimicrobial resistance in ICU. Intensive Care Med 2016 Jun;42(6):955-958.
- 24. MacVane SH. Antimicrobial resistance in the intensive care unit: a focus on gram-negative bacterial infections. J Intensive Care Med 2017 Jan;32(1):25-37.
- 25. Harris PN, Peleg AY, Iredell J, Ingram PR, Miyakis S, Stewardson AJ, et al. Meropenem versus piperacillin-tazobactam for definitive treatment of bloodstream infections due to ceftriaxone non-susceptible Escherichia coli and Klebsiella spp (the MERINO trial): study protocol for a randomised controlled trial. Trials 2015 Jan;16:24.
- 26. Livermore DM, Mushtaq S, Meunier D, Hopkins KL, Hill R, Adkin R, et al; BSAC Resistance surveillance Standing Committee. Activity of ceftolozane/tazobactam against surveillance and ‘problem’ Enterobacteriaceae, Pseudomonas aeruginosa and non-fermenters from the British Isles. J Antimicrob Chemother 2017 Aug;72(8):2278-2289.
- 27. Khan FY, Elshafie SS, Almaslamani M, Abu-Khattab M, El Hiday AH, Errayes M, et al. Epidemiology of bacteraemia in Hamad general hospital, Qatar: a one year hospital-based study. Travel Med Infect Dis 2010 Nov;8(6):377-387.
- 28. Shaikh N, Momin U, Atef Shible A, Al-Musalmani M, Ansari A. Community acquired urosepsis: a surgical intensive care experience. Qatar Med J 2020 Apr;2020(1):8.
- 29. Balkhy HH, Assiri AM, Mousa HA, Al-Abri SS, Al-Katheeri H, Alansari H, et al; at the workshop. The strategic plan for combating antimicrobial resistance in Gulf Cooperation Council States. J Infect Public Health 2016 Jul-Aug;9(4):375-385.
- 30. Aly M, Balkhy HH. The prevalence of antimicrobial resistance in clinical isolates from Gulf Corporation Council countries. Antimicrob Resist Infect Control 2012 Jul;1(1):26.
- 31. Khanfar H. Molecular epidemiological study of extended spectrum beta-lactamase (ESBL) producing bacteria from a hospital within Saudi Arabia. University of Portsmouth, 2017.
- 32. Perez-Lopez A, Sundararaju S, Al-Mana H, Tsui KM, Hasan MR, Suleiman M, et al. Molecular characterization of extended-spectrum β-lactamase-producing escherichia coli and Klebsiella pneumoniae among the pediatric population in Qatar. Front Microbiol 2020 Nov;11:581711.
- 33. Abid FB, Tsui CK, Doi Y, Deshmukh A, McElheny CL, Bachman WC, et al. Molecular characterization of clinical carbapenem-resistant Enterobacterales from Qatar. Eur J Clin Microbiol Infect Dis 2021 Aug;40(8):1779-1785.
- 34. Zalas-Więcek P, Gospodarek-Komkowska E. Antimicrobial susceptibility of multi-drug and extensively-drug-resistant Escherichia coli to ceftolozane-tazobactam and ceftazidime-avibactam: an in vitro study. Postepy Hig Med Dosw 2020;74:77-83.
- 35. Ortiz de la Rosa J-M, Nordmann P, Poirel L. ESBLs and resistance to ceftazidime/avibactam and ceftolozane/tazobactam combinations in Escherichia coli and pseudomonas aeruginosa. J Antimicrob Chemother 2019 Jul;74(7):1934-1939.
- 36. Lahiri SD, Alm RA. Identification of novel VEB β-lactamase enzymes and their impact on avibactam inhibition. Antimicrob Agents Chemother 2016 Apr;60(5):3183-3186.
- 37. Barnes MD, Taracila MA, Rutter JD, Bethel CR, Galdadas I, Hujer AM, et al. Deciphering the evolution of cephalosporin resistance to ceftolozane-tazobactam in pseudomonas aeruginosa. mBio 2018 Dec;9(6):e02085-18.
- 38. Arca-Suárez J, Lasarte-Monterrubio C, Rodiño-Janeiro B-K, Cabot G, Vázquez-Ucha JC, Rodríguez-Iglesias M, et al. Molecular mechanisms driving the in vivo development of OXA-10-mediated resistance to ceftolozane/tazobactam and ceftazidime/avibactam during treatment of XDR Pseudomonas aeruginosa infections. J Antimicrob Chemother 2021 Jan;76(1):91-100.
- 39. Cultrera R, Libanore M, Barozzi A, d’Anchera E, Romanini L, Fabbian F, et al. Ceftolozane/tazobactam and ceftazidime/avibactam for multidrug-resistant gram-negative infections in immunocompetent patients: a single-center retrospective study. Antibiotics (Basel) 2020 Sep;9(10):640.
- 40. Evans SR, Tran TT, Hujer AM, Hill CB, Hujer KM, Mediavilla JR, et al; Antibacterial resistance leadership group (ARLG). Rapid molecular diagnostics to inform empiric use of ceftazidime/avibactam and ceftolozane/tazobactam against pseudomonas aeruginosa: PRIMERS IV. Clin Infect Dis 2019 May;68(11):1823-1830.
Table A1: Microbiological characteristics, molecular characterization, and susceptibility testing results for 109 ESBL-producing Enterobacteriaceae isolates.
|
1
|
Nov-12
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Sputum
|
Positive
|
Negative
|
Negative
|
Positive
|
0.25
|
0.75
|
|
2
|
Nov-12
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Urine
|
Positive
|
Positive
|
Positive
|
Positive
|
0.25
|
0.50
|
|
3
|
Nov-12
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Sputum
|
Positive
|
Positive
|
Positive
|
Positive
|
0.75
|
1.00
|
|
4
|
Nov-12
|
Escherichia coli
|
SICU
|
Wound Swab
|
Positive
|
Negative
|
Negative
|
Positive
|
0.12
|
0.38
|
|
5
|
Nov-12
|
Serratia marcescens
|
MICU
|
Blood
|
Positive
|
Negative
|
Negative
|
Negative
|
0.12
|
0.19
|
|
6
|
Nov-12
|
Klebsiella pneumoniae ssp pneumoniae
|
PICU
|
Endotracheal Tube Secretion
|
Positive
|
Positive
|
Negative
|
Negative
|
0.19
|
0.38
|
|
7
|
Nov-12
|
Klebsiella pneumoniae ssp pneumoniae
|
NICU
|
Peritoneal fluid
|
Negative +AmpC
|
Negative
|
Positive
|
Positive
|
0.19
|
0.25
|
|
8
|
Nov-12
|
Escherichia coli
|
MICU
|
Urine
|
Negative +AmpC
|
Negative
|
Positive
|
Negative
|
0.50
|
2.00
|
|
9
|
Nov-12
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Blood
|
Positive
|
Positive
|
Negative
|
Negative
|
0.75
|
1.50
|
|
10
|
Nov-12
|
Klebsiella pneumoniae ssp pneumoniae
|
PICU
|
Endotracheal Tube Secretion
|
Positive
|
Positive
|
Positive
|
Positive
|
0.19
|
0.38
|
|
11
|
Dec-12
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Endotracheal Tube Secretion
|
Positive
|
Positive
|
Positive
|
Positive
|
0.19
|
0.38
|
|
12
|
Dec-12
|
Klebsiella oxytoca
|
NICU
|
Tracheostomy Site Swab
|
Positive
|
Negative
|
Negative
|
Negative
|
0.12
|
0.38
|
|
13
|
Dec-12
|
Klebsiella pneumoniae ssp pneumoniae
|
PICU
|
Urine
|
Positive
|
Positive
|
Positive
|
Positive
|
0.25
|
0.50
|
|
14
|
Dec-12
|
Escherichia coli
|
NICU
|
Conjunctival Swab
|
Positive
|
Negative
|
Negative
|
Positive
|
0.09
|
0.38
|
|
15
|
Dec-12
|
Enterobacter aerogenes
|
MICU
|
Blood
|
Positive
|
Positive
|
Negative
|
Negative
|
0.19
|
0.50
|
|
16
|
Dec-12
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
Endotracheal Tube Secretion
|
Positive
|
Positive
|
Positive
|
Positive
|
0.19
|
0.38
|
|
17
|
Jan-13
|
Escherichia coli
|
MICU
|
Urine
|
Negative
|
Negative
|
Negative
|
Negative
|
0.38
|
0.50
|
|
18
|
Jan-13
|
Escherichia coli
|
SICU
|
Blood
|
Positive
|
Negative
|
Positive
|
Negative
|
0.25
|
0.38
|
|
20
|
Jan-13
|
Enterobacter cloacae
|
TICU
|
Blood
|
Negative
|
Negative
|
Negative
|
Negative
|
0.12
|
0.25
|
|
21
|
Jan-13
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Sputum
|
Positive
|
Positive
|
Positive
|
Positive
|
0.25
|
0.38
|
|
22
|
Jan-13
|
Escherichia coli
|
MICU
|
Blood
|
Negative +AmpC
|
Negative
|
Positive
|
Negative
|
0.38
|
1.50
|
|
23
|
Jan-13
|
Proteus penneri
|
MICU
|
Blood
|
Positive
|
Negative
|
Negative
|
Negative
|
0.047
|
1.00
|
|
24
|
Jan-13
|
Escherichia coli
|
PICU
|
Urine
|
Positive
|
Negative
|
Negative
|
Positive
|
0.12
|
0.38
|
|
25
|
Jan-13
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
Endotracheal Tube Secretion
|
Positive
|
Positive
|
Negative
|
Negative
|
0.25
|
0.38
|
|
26
|
Jan-13
|
Klebsiella pneumoniae ssp pneumoniae
|
TICU
|
Urine
|
Positive
|
Positive
|
Positive
|
Positive
|
0.25
|
0.75
|
|
27
|
Jan-13
|
Citrobacter braakii
|
TICU
|
Blood
|
Negative
|
Negative
|
Negative
|
Negative
|
0.25
|
0.75
|
|
28
|
Jan-13
|
Escherichia coli
|
TICU
|
Sputum
|
Positive
|
Negative
|
Negative
|
Positive
|
0.94
|
0.38
|
|
29
|
Jan-13
|
Escherichia coli
|
PICU
|
Urine
|
Positive
|
Negative
|
Positive
|
Positive
|
0.12
|
0.38
|
|
30
|
Feb-13
|
Serratia marcescens
|
MICU
|
Sputum
|
Negative
|
Negative
|
Negative
|
Negative
|
0.02
|
0.19
|
|
31
|
Feb-13
|
Escherichia coli
|
MICU
|
Urine
|
Positive
|
Negative
|
Positive
|
Positive
|
0.06
|
0.25
|
|
32
|
Feb-13
|
Escherichia coli
|
MICU
|
Blood
|
Positive
|
Negative
|
Negative
|
Positive
|
0.12
|
0.50
|
|
33
|
Feb-13
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
Tracheal Aspirate
|
Positive
|
Positive
|
Positive
|
Positive
|
0.19
|
0.38
|
|
34
|
Feb-13
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Urine
|
Positive
|
Positive
|
Negative
|
Positive
|
0.09
|
0.38
|
|
35
|
Feb-13
|
Escherichia coli
|
MICU
|
Blood
|
Positive
|
Negative
|
Negative
|
Positive
|
0.19
|
0.38
|
|
36
|
Mar-13
|
Escherichia coli
|
TICU
|
Ascitic Fluid
|
Positive
|
Negative
|
Negative
|
Positive
|
0.12
|
0.38
|
|
37
|
Mar-13
|
Klebsiella pneumoniae ssp pneumoniae
|
TICU
|
Sputum
|
Positive
|
Positive
|
Negative
|
Negative
|
0.25
|
0.38
|
|
38
|
Mar-13
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Urine
|
Positive
|
Negative
|
Negative
|
Positive
|
0.38
|
0.38
|
|
39
|
Mar-13
|
Klebsiella pneumoniae ssp pneumoniae
|
NICU
|
Blood
|
Positive
|
Positive
|
Negative
|
Positive
|
0.25
|
0.38
|
|
40
|
Mar-13
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Blood
|
Positive
|
Positive
|
Negative
|
Positive
|
0.25
|
0.38
|
|
42
|
Mar-13
|
Escherichia coli
|
SICU
|
Peritoneal fluid
|
Negative + AmpC
|
Negative
|
Negative
|
Positive
|
0.19
|
0.75
|
|
43
|
Mar-13
|
Escherichia coli
|
SICU
|
Urine
|
Positive
|
Positive
|
Negative
|
Positive
|
0.12
|
0.38
|
|
44
|
Mar-13
|
Klebsiella pneumoniae ssp pneumoniae
|
PICU
|
Urine
|
Positive
|
Positive
|
Positive
|
Positive
|
0.19
|
0.5
|
|
45
|
Mar-13
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
Sputum
|
Positive
|
Positive
|
Positive
|
Positive
|
0.38
|
1.00
|
|
46
|
Mar-13
|
Klebsiella pneumoniae ssp ozaenae
|
MICU
|
Sputum
|
Positive
|
Negative
|
Negative
|
Negative
|
0.09
|
0.25
|
|
47
|
Mar-13
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
Tracheal Aspirate
|
Positive
|
Negative
|
Positive
|
Positive
|
0.12
|
0.25
|
|
48
|
Mar-13
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
Urine
|
Positive
|
Positive
|
Positive
|
Positive
|
0.38
|
1.50
|
|
49
|
Mar-13
|
Escherichia coli
|
TICU
|
Urine
|
Positive
|
Negative
|
Positive
|
Positive
|
0.09
|
0.38
|
|
50
|
Apr-13
|
Klebsiella pneumoniae ssp pneumoniae
|
TICU
|
J VAC Fluid
|
Positive
|
Negative
|
Positive
|
Positive
|
0.19
|
0.5
|
|
51
|
Apr-13
|
Escherichia coli
|
TICU
|
J VAC Fluid
|
Positive
|
Negative
|
Negative
|
Positive
|
0.09
|
0.75
|
|
52
|
Apr-13
|
Escherichia coli
|
PICU
|
Urine
|
Positive
|
Negative
|
Negative
|
Positive
|
0.12
|
0.38
|
|
53
|
Apr-13
|
Klebsiella pneumoniae ssp pneumoniae
|
PICU
|
Endotracheal Tube Secretion
|
Positive
|
Positive
|
Positive
|
Positive
|
0.19
|
0.75
|
|
54
|
Apr-13
|
Enterobacter aerogenes
|
MICU
|
Tracheal Aspirate
|
Positive
|
Positive
|
Positive
|
Positive
|
0.25
|
0.38
|
|
55
|
Apr-13
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Blood
|
Positive
|
Positive
|
Negative
|
Negative
|
0.75
|
1.00
|
|
56
|
Apr-13
|
Klebsiella pneumoniae ssp pneumoniae
|
PICU
|
Endotracheal Tube Secretion
|
Positive
|
Positive
|
Negative
|
Negative
|
0.25
|
0.38
|
|
57
|
Apr-13
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Blood
|
Positive
|
Positive
|
Negative
|
Positive
|
0.19
|
0.38
|
|
58
|
Apr-13
|
Escherichia coli
|
SICU
|
Wound Swab
|
Positive
|
Negative
|
Negative
|
Positive
|
0.12
|
0.50
|
|
59
|
Apr-13
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Sputum
|
Positive
|
Positive
|
Negative
|
Negative
|
0.25
|
0.38
|
|
60
|
Apr-13
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Blood
|
Positive
|
Positive
|
Negative
|
Negative
|
0.19
|
0.25
|
|
62
|
Apr-13
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
Endotracheal Tube Secretion
|
Positive
|
Positive
|
Negative
|
Negative
|
0.25
|
0.25
|
|
63
|
May-13
|
Citrobacter freundii
|
TICU
|
Blood
|
Negative
|
Negative
|
Negative
|
Negative
|
0.06
|
0.38
|
|
64
|
May-13
|
Escherichia coli
|
MICU
|
Urine
|
Positive
|
Negative
|
Negative
|
Positive
|
0.06
|
0.25
|
|
65
|
May-13
|
Klebsiella pneumoniae ssp pneumoniae
|
PICU
|
Tracheostomy Site Swab
|
Positive
|
Positive
|
Positive
|
Positive
|
0.25
|
0.75
|
|
66
|
May-13
|
Klebsiella pneumoniae ssp pneumoniae
|
NICU
|
Endotracheal Tube Secretion
|
Positive
|
Positive
|
Positive
|
Positive
|
0.25
|
0.38
|
|
67
|
May-13
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Sputum
|
Positive
|
Positive
|
Negative
|
Negative
|
0.19
|
0.38
|
|
68
|
May-13
|
Klebsiella pneumoniae ssp pneumoniae
|
TICU
|
Blood
|
Positive
|
Positive
|
Negative
|
Positive
|
0.19
|
0.75
|
|
69
|
May-13
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
Sputum
|
Positive
|
Positive
|
Negative
|
Negative
|
0.38
|
0.38
|
|
70
|
May-13
|
Enterobacter cloacae
|
SICU
|
Sputum
|
Positive
|
Positive
|
Negative
|
Negative
|
0.19
|
0.19
|
|
71
|
Jun-13
|
Klebsiella pneumoniae ssp pneumoniae
|
TICU
|
Wound Swab
|
Positive
|
Positive
|
Negative
|
Positive
|
0.38
|
0.38
|
|
72
|
Jun-13
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Blood
|
Positive
|
Positive
|
Negative
|
Negative
|
0.19
|
0.38
|
|
73
|
Jun-13
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Blood
|
Positive
|
Positive
|
Positive
|
Positive
|
0.19
|
1.00
|
|
74
|
Jun-13
|
Escherichia coli
|
PICU
|
Endotracheal Tube Secretion
|
Positive
|
Negative
|
Negative
|
Positive
|
0.12
|
0.38
|
|
75
|
Jun-13
|
Escherichia coli
|
PICU
|
Urine
|
Positive
|
Negative
|
Negative
|
Positive
|
0.12
|
0.38
|
|
76
|
Jun-13
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Blood
|
Positive
|
Positive
|
Positive
|
Positive
|
0.38
|
1.50
|
|
77
|
Jul-13
|
Klebsiella pneumoniae ssp pneumoniae
|
NICU
|
Blood
|
Positive
|
Positive
|
Negative
|
Negative
|
0.38
|
0.38
|
|
78
|
Jul-13
|
Escherichia coli
|
NICU
|
Blood
|
Positive
|
Negative
|
Negative
|
Negative
|
0.12
|
0.38
|
|
79
|
Jul-13
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
Sputum
|
Positive
|
Positive
|
Negative
|
Positive
|
0.25
|
0.75
|
|
80
|
Jul-13
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Endotracheal Tube Secretion
|
Positive
|
Positive
|
Positive
|
Positive
|
0.19
|
0.38
|
|
82
|
Jul-13
|
Escherichia coli
|
SICU
|
Urine
|
Positive
|
Negative
|
Positive
|
Negative
|
0.25
|
0.50
|
|
83
|
Jul-13
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
BAL
|
Positive
|
Positive
|
Negative
|
Positive
|
0.25
|
0.75
|
|
84
|
Jul-13
|
Escherichia coli
|
MICU
|
Urine
|
Positive
|
Negative
|
Negative
|
Negative
|
0.09
|
0.25
|
|
85
|
Jul-13
|
Citrobacter amalonaticus
|
PICU
|
Urine
|
Positive
|
Positive
|
Negative
|
Negative
|
0.12
|
0.19
|
|
86
|
Jul-13
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
Sputum
|
Positive
|
Positive
|
Positive
|
Positive
|
0.25
|
0.50
|
|
87
|
Jul-13
|
Enterobacter cloacae
|
NICU
|
Eye Swab
|
Negative + AmpC
|
Negative
|
Negative
|
Negative
|
0.09
|
0.25
|
|
88
|
Jul-13
|
Enterobacter cloacae
|
TICU
|
Blood
|
Negative + AmpC
|
Negative
|
Negative
|
Negative
|
0.09
|
0.25
|
|
89
|
Jul-13
|
Enterobacter aerogenes
|
TICU
|
Blood
|
Positive
|
Positive
|
Negative
|
Negative
|
0.09
|
0.38
|
|
90
|
Aug-13
|
Escherichia coli
|
MICU
|
Ascitic fluid
|
Positive
|
Negative
|
Positive
|
Positive
|
0.12
|
0.25
|
|
91
|
Aug-13
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
Urine
|
Positive
|
Positive
|
Negative
|
Positive
|
0.38
|
0.75
|
|
92
|
Aug-13
|
Enterobacter aerogenes
|
TICU
|
Sputum
|
Positive
|
Positive
|
Negative
|
Negative
|
0.25
|
0.38
|
|
93
|
Aug-13
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
Sputum
|
Positive
|
Positive
|
Positive
|
Positive
|
0.19
|
0.75
|
|
94
|
Sep-13
|
Escherichia coli
|
NICU
|
Blood
|
Positive
|
Negative
|
Negative
|
Positive
|
0.19
|
0.75
|
|
95
|
Sep-13
|
Klebsiella pneumoniae ssp pneumoniae
|
PICU
|
Urine
|
Positive
|
Positive
|
Positive
|
Positive
|
0.25
|
0.5
|
|
96
|
Sep-13
|
Escherichia coli
|
MICU
|
Urine
|
Positive
|
Negative
|
Positive
|
Positive
|
0.09
|
0.38
|
|
97
|
Sep-13
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
Urine
|
Positive
|
Positive
|
Positive
|
Positive
|
0.25
|
0.75
|
|
98
|
Sep-13
|
Escherichia coli
|
SICU
|
Sputum
|
Positive
|
Negative
|
Negative
|
Positive
|
0.12
|
0.50
|
|
99
|
Sep-13
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
Sputum
|
Positive
|
Positive
|
Positive
|
Positive
|
0.25
|
0.75
|
|
100
|
Sep-13
|
Klebsiella pneumoniae ssp pneumoniae
|
PICU
|
Blood
|
Positive
|
Negative
|
Positive
|
Positive
|
0.12
|
0.25
|
|
101
|
Sep-13
|
Escherichia coli
|
PICU
|
Urine
|
Positive
|
Negative
|
Negative
|
Positive
|
0.06
|
0.25
|
|
102
|
Sep-13
|
Klebsiella pneumoniae ssp pneumoniae
|
MICU
|
Sputum
|
Positive
|
Positive
|
Positive
|
Positive
|
0.19
|
0.38
|
|
103
|
Sep-13
|
Klebsiella pneumoniae ssp pneumoniae
|
NICU
|
Central line Tip
|
Positive
|
Positive
|
Negative
|
Positive
|
0.25
|
0.50
|
|
104
|
Sep-13
|
Escherichia coli
|
TICU
|
Sputum
|
Positive
|
Negative
|
Positive
|
Positive
|
0.19
|
0.25
|
|
106
|
Oct-13
|
Escherichia coli
|
SICU
|
Sputum
|
Positive
|
Positive
|
Positive
|
Negative
|
0.06
|
0.19
|
|
107
|
Oct-13
|
Escherichia coli
|
SICU
|
Urine
|
Positive
|
Negative
|
Negative
|
Positive
|
0.09
|
0.25
|
|
108
|
Oct-13
|
Klebsiella pneumoniae ssp pneumoniae
|
SICU
|
Blood
|
Positive
|
Positive
|
Positive
|
Positive
|
0.38
|
1.00
|
ESBL: extended-spectrum β-lactamase; MIC: minimum inhibitory concentration; White: susceptible; grey: non-susceptible, susceptibility was reported according to Clinical Laboratory Standards Institute (CLSI) breakpoints (Clinical Laboratory Standards Institute, 2020). m: month; y: year; MICU: Medical Intensive Care Unit; NICU: Neonatal Intensive Care Unit; PICU: Pediatric Intensive Care Unit; SICU: Surgical Intensive Care Unit; TICU: Trauma Intensive Care Unit; CZA: ceftazidime-avibactam; C/T: ceftolozane-tazobactam.