Depressive disorders are characterized by chronic sadness and feelings of emptiness or irritable mood, along with changes in somatization and cognitions.1 Depressive disorders are more common in women than men. The various risk factors leading to depression are environmental and genetic factors. Environmental factors, particularly abuse and neglect in childhood, are risk factors for various psychiatric symptoms and disorders, particularly mood and anxiety disorders.2 However, 30–40% of the risk for developing depression is genetically determined.3
No nationwide data is currently available on the prevalence of depression in the UAE, both for citizens and residents. Most research conducted has focused on studying depression in selected small groups of people. For example, the prevalence rate of depression was 25.1% among male migrant workers living in the UAE,4 6–22% among medical residents in the country,5 10% (postpartum depression) among women in Sharjah,6 17.6% among patients with multiple sclerosis,7 63.3% among resident doctors working for the Dubai Health Authority,8 and 22.2% among university students.9 Monsef et al,8 also reported a prevalence rate of 57.4% for anxiety among resident doctors. A recent systematic review found that among the residing population of the UAE, female sex, family history of chronic diseases, low socioeconomic status, lack of social support, and stressful life events are significant risk factors in developing depression.10
Many researchers have focused on genetic risk factors associated with various psychiatric conditions, including depression. Among these genes, serotonin transporter gene promoter polymorphism (SLC6A4) was strongly associated with neuroticism as a personality trait characterized by depression and anxiety and other negative emotions.11,12 Hettema et al,13 highlighted the role of the PPARGC1A gene as a potential susceptibility gene for anxiety-related disorders. A comprehensive genetic study found that the CAMKMT gene has a significant association with anxiety phenotypes in a large sample of patients with European ancestry.14
The level of the monoamine neurotransmitter in the brain is to a large extent regulated by monoamine oxidase A (MAOA). MAOA inhibitors have been effectively used for the treatment of depression.15 It has been found that MAOA upstream variable-number tandem repeat (u-VNTR) alleles portrayed a reduction in the amygdala-prefrontal cortex connectivity, which was significantly associated with acute major depression.15 A similar significant association between MAOA u-VNTR and major depressive disorder, especially among females, was also found in other studies.16,17
The 11 beta-hydroxysteroid dehydrogenase type 1 enzyme, which is associated with the HSD11B1 gene, is responsible for converting cortisone into cortisol.18 The amount of cortisol in body fluids such as blood, saliva, or urine is often used to measure stress in a person. HSD11B1 has previously been linked to health problems such as insulin resistance, obesity, metabolic syndrome and diabetes, and also positively associated with polycystic ovary syndrome.19
We sought to study the expression of various genes (PPARGC1A, CAMKMT, SLC6A4, MAOA, and HSD11B1) associated with depression to identify genetic risk factors associated with its development in the UAE population. To the best of our knowledge, this study is the first to study the selected genes’ association with depression in the residing UAE population.
Method
Participants for the current study were recruited from Al Ain Hospital through random sampling and from UAE University through convenience sampling methods. All of the recruited participants resided in the UAE and were from various nationalities. Patients and controls were asked to read and sign the consent form before participation in the study. They were also required to complete the Patient Health Questionnaire-9 (PHQ-9) and the Beck Depression Inventory-II (BDI-II) before collecting blood samples. The BDI-II and PHQ-9 were chosen for this study because their Arabic versions have been validated and are reliable.20,21
We recruited 29 patients aged 18–65 years (case group) from the outpatient psychiatric clinic at Al Ain Hospital, and 30 healthy controls from UAE University. We have completed study from September 1, 2018 to May 30, 2019. The mean age of the depressed group was 40.3 years and 26.5 years in the control group. Participants in the depressed group were included if they met the criteria for diagnosis of a mood disorder by a licensed psychiatrist according to the International Classification of Disease 10th revision. Participants in this group were excluded if they had (1) comorbid psychotic features, (2) comorbid psychiatric disorders such as schizophrenia or personality disorders, or (3) current substance abuse. All of this information was gathered through a semi-structured interview by the psychiatrist giving the diagnoses. Participants in the control group were excluded if they scored above 13 on the BDI-II or above 9 on the PHQ-9. In addition, participants were matched on the gender and nationality variables. A minimum of 30 participants per group were targeted to gain statistically significant results. We recruited 32 participants in the case group and 33 participants in the control group. However, due to the low quality of RNA found in the blood samples of six participants, they were excluded from the study. As a result, 29 participants were included in the case group and 30 in the control group.
The study was approved by the ethical committees of Al Ain Hospital and United Arab Emirates University.
The PHQ-9 is the depression module of the PHQ questionnaire. It has nine self-administered questions based on the diagnostic criteria of depression according to the diagnostic and statistical manual of mental disorders-IV. Each question is scored on a four-point scale from 0 (not at all) to 3 (nearly every day). Higher scores reflect greater severity.22 Good discriminant validity for the Arabic version was confirmed and reliability using Cronbach’s alpha was 0.857.21
The BDI-II is a 21-item self-reported multiple-choice inventory that takes approximately 10 min to complete. It is a widely used tool to indicate the severity of depression. It is available in several languages and is appropriate for individuals aged 13–80 years old. The items on this scale are rated on a four-point scale from 0 (symptoms not present) to 3 (symptoms strongly present), with a maximum total score of 63.23 Scores between 0–13 indicate normal-to-minimal depression, between 14–19 indicate mild depression, between 20–28 indicate moderate depression, and between 29–63 indicate severe depression. The Arabic version of the BDI-II has a high degree of validity and reliability.20
PHQ-9 was used in this study along with the BDI-II as a screening tool for depression as it has been found that the BDI-II tends to characterize a larger proportion of participants with severe depression than the PHQ-9.24 Thus, BDI-II was used to assess the severity of the depressive symptoms.
Venous blood samples were obtained from all subjects (depressed and control) in a PAXgene collection tube provided by Qiagen. These samples were stored at -20 °C. RNA extraction from blood samples were extracted using the PAXgene Blood RNA Kit provided by PreAnalytiX (Qiagen; Ref #762164). RNA purity was measured using a spectrophotometer.
Primers for all the genes were selected using the TaqMan assays and arrays tool provided by Thermo Fisher. cDNA synthesis was performed using the Thermo Fisher Scientific High-Capacity cDNA Reverse Transcription Kit (Ref #4368814). Reverse transcription polymerase chain reaction (RT-PCR) was carried out using the Thermal Cycler provided by Thermo Fisher Scientific. The QuantStudio Real-Time PCR by Thermo Fisher Scientific was used to run the qRT-PCR. A mixture of 10 μL was prepared for each reaction using 8 μL of master mix containing gene primers and 2 μL of 10 ng cDNA for each sample and loaded on a 96-well plate. The following conditions were set up for the plate: UNG incubation at 50 °C for two min, polymerase activation at 95 °C for two min, PCR denature cycle at 95 °C for three sec, and PCR anneal cycle at 60 °C for 30 sec.
We used SPSS Statistics (IBM Corp. Released 2012. IBM SPSS Statistics for Windows, Version 21.0. Armonk, NY: IBM Corp.) and QuantStudio Design and Analysis Software (version 1.3.1) for comparative and descriptive analysis of the data.
Results
We targeted two main groups of UAE residents. The case group included 29 patients with depression and the control group included 30 non-depressed patients [Table 1]. We used the independent-samples t-test to examine the baseline level of depression for both groups [Table 2].
Gene expression analysis was carried using the QuantStudio Design and Analysis Software. The authors targeted the five genes’ expression levels compared to a reference level of 1.0 [Figure 1]. The independent samples t-test was used to check the significance of the difference between the expression levels of the genes [Table 3]. No significant difference in the genes’ expression (CAMKMT, SLC6A4, and HSD11B1) were found for nationality or gender within each group, so the data were pooled. The PPARGC1A gene was the most significantly downregulated, whereas MAOA expression was upregulated in the case group compared with the control group [Table 3]. Independent samples t-test was also run for the MAOA gene between both genders, but no significant difference was seen (p = 0.470) [Table 4].
Table 1: Descriptive statistics for total number and percentage of participants in gender and nationality variables.
|
Gender |
|
|
|
|
|
Male |
15 |
51.7 |
12 |
40.0 |
|
Female |
14 |
48.3 |
18 |
60.0 |
|
Total |
29 |
100 |
30 |
100 |
|
Nationality |
|
|
|
|
|
Emirati |
12 |
41.4 |
15 |
50.0 |
|
Non-Emirati |
17 |
58.6 |
15 |
50.0 |
Table 2: Independent samples t-test for baseline depression.
|
BDI |
|
|
|
|
|
|
|
Case |
29 |
31.9 |
10.7 |
12.73 |
57 |
< 0.001 |
|
Control |
30 |
5.5 |
3.5 |
|
|
|
|
PHQ-9 |
|
|
|
|
|
|
|
Case |
29 |
14.3 |
4.6 |
13.32 |
57 |
< 0.001 |
SD: standard deviation; df: degree of freedom; BDI: Beck Depression Inventory; PHQ-9: Patient Health Questionnaire-9.
Mean BDI and PHQ-9 scores among the case and control groups. The level of depression among the case group was significantly higher than the control group on both scales.
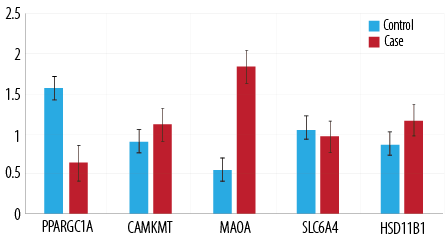
Figure 1: Gene expression in the case and control patient groups.
Table 3: Independent samples t-test for gene expression.
|
PPARGC1A |
Case |
29 |
0.4 |
0.3 |
0.4 |
2.39 |
57 |
0.020 |
|
Control |
30 |
0.8 |
0.7 |
|
CAMKMT |
Case |
29 |
1.3 |
0.5 |
-0.2 |
-1.46 |
57 |
0.150 |
|
Control |
30 |
1.1 |
0.3 |
|
MAOA |
Case |
29 |
2.0 |
2.2 |
-1.0 |
-1.83 |
57 |
0.070 |
|
Control |
30 |
1.0 |
1.8 |
|
SLC6A4 |
Case |
29 |
1.3 |
0.9 |
0.0 |
0.32 |
57 |
0.750 |
|
Control |
30 |
1.3 |
0.8 |
|
Case |
29 |
1.8 |
1.1 |
SD: standard deviation; df: degree of freedom.
Mean expression of each gene among the case and control groups. We found a significant difference in the expression of PPARGC1A at p < 0.050 between the case and control groups. However, there was no significant difference in expression of CAMKMT, MAOA, SLC6A4, and HSD11B1 between the groups.
Mean expression of MAOA among the case and control groups. There is no significant difference in the expression of MAOA between the groups.
Table 4: Independent samples t-test for MAOA expression gender comparison.
SD: standard deviation; df: degree of freedom.
Discussion
Only the PPARGC1A gene showed a significant difference between the two groups (p < 0.050). The PPARGC1A gene has previously been shown to be related to anxious phenotypes in genome-wide association analysis.13 Our results revealed that the expression of PPARGC1A is significantly higher in the control group than in the group of patients with depressive disorder. Our results highlight the importance of PPARGCIA gene in association with depression in the residing UAE population. Furthermore, the results indicate a possible protective role that the gene may play against developing depressive disorders.
PPARGC1A is a transcriptional coactivator for steroid and nuclear receptors. It plays an important role in energy metabolism and is involved in the cellular response to oxidative stress and negative regulation of neuronal death. It could be the case that higher expression of the gene protects individuals from developing a depressive disorder and a decrease in the expression makes them more vulnerable to it. Interestingly, PPARGC1A is responsive to several forms of environmental stressors, including nutritional status and temperature,25,26 making it a good candidate for evidence for the epigenetics model. Previous research has found elevated levels of this gene to protect neural cells against apoptosis caused by oxidative stress27 and improve neurological symptoms.28 Low levels of PPARGC1A have also been shown to be associated with Alzheimer’s disease and memory loss.29 Since depression is known to precede clinical diagnosis of Alzheimer’s disease and have a prevalence of up to 50% in these patients, it is important to study the underlying mechanism of PPARGC1A to better understand its role against developing depressive disorders.
The difference in expression for the other genes was not significant. This contradicts previous research linking these genes to depressive phenotypes.11,12,13 Additionally, even though the expression of the MAOA gene has the greatest mean difference (-1.0) between the two groups, the difference is not statistically significant (p = 0.070). This could be due to the large standard deviation (2.2 and 1.8) between the individual values among the samples and the highest standard error mean (0.5) among all the genes. HSD11B1 was linked to anxiety phenotypes in a preliminary study.30 No confirmatory analysis has been conducted to confirm this linkage. The mean difference of HSD11B1 expression between the two groups is -0.4 and was not statistically significant (p = 0.100). This provides novel information that this gene is not associated with depressive disorders. However, it does not reject the results obtained from the preliminary study as no baseline of anxiety levels was collected from the participants.
This research has several limitations. Firstly, the relatively small sample size may have played a role in the overall results. Secondly, the two comparison groups were not matched for age, as the control group was much younger, comprising mainly university students. Lastly, the participants for the study originated from different countries. Having homogeneity in ethnicity and nationality could have provided much stronger evidence for genetic associations.
The current study is one of the very few studies conducted on genetic association with depression in the region. No such previous study has been published in the country to the best of our knowledge.
Further research is recommended to confirm the results obtained in the current study. Future studies can focus on the pathways involved in the functioning of PPARGC1A to understand its role in depressive disorders. It is also recommended to explore its possible association with the upregulation of key neurotransmitters associated with depression, such as serotonin and dopamine. Further research is also recommended to keep the nationality variable constant. A larger sample size can also add significance to future research.
Conclusion
Our study focused on exploring the expression of genetic biomarkers among patients with depression by comparing them to a non-depressed control group. Five genes were selected for analysis. The genes associated with anxiety were selected due to high comorbidity between depressive and anxiety disorders. Studying these genes among a depressed population would provide insight into the prevalence of anxiety genes among depressed individuals or the role of such genes in mood disorders. Among the five genes analyzed, only the PPARGC1A gene was seen to have a statistically significant difference in the expression levels between the two groups. This result highlights the role that PPARGC1A may play in protecting against depressive disorders. Contrary to previous research, no other significant differences among gene expressions between the two groups were observed in the current study.
Disclosure
The authors declared no conflicts of interest. No funding was received for this study.
references
- 1. American Psychiatric Association. American Psychiatric Association DSM-5 Diagnostic and statistical manual of mental disorders: DSM-5. Washington, D.C.: American Psychiatric Association, 2013.
- 2. Heim C, Shugart M, Craighead WE, Nemeroff CB. Neurobiological and psychiatric consequences of child abuse and neglect. Dev Psychobiol 2010 Nov;52(7):671-690.
- 3. Heim C, Binder EB. Current research trends in early life stress and depression: review of human studies on sensitive periods, gene-environment interactions, and epigenetics. Exp Neurol 2012 Jan;233(1):102-111.
- 4. Al-Maskari F, Shah SM, Al-Sharhan R, Al-Haj E, Al-Kaabi K, Khonji D, et al. Prevalence of depression and suicidal behaviors among male migrant workers in United Arab Emirates. J Immigr Minor Health 2011 Dec;13(6):1027-1032.
- 5. Abdulrahman M, Nair SC, Farooq MM, Al Kharmiri A, Al Marzooqi F, Carrick FR. Burnout and depression among medical residents in the United Arab Emirates: a multicenter study. J Family Med Prim Care 2018 Mar-Apr;7(2):435-441.
- 6. Hamdan A, Tamim H. Psychosocial risk and protective factors for postpartum depression in the United Arab Emirates. Arch Womens Ment Health 2011 Apr;14(2):125-133.
- 7. Alsaadi T, Hammasi KE, Shahrour TM, Shakra M, Turkawi L, Nasreddine W, et al. Depression and anxiety as determinants of health-related quality of life in patients with multiple sclerosis - United Arab Emirates. Neurol Int 2017 Dec;9(4):7343.
- 8. Monsef NA, Hajaj KE, Basti AK, Marzouqi EA, Faisal WA, Hussein H, et al. Perceived depression, anxiety and stress among Dubai Health authority residents, Dubai, UAE. American Journal of Psychology and Cognitive Science 2015;1(3):75-82.
- 9. Mellal AA, Albluwe T, Al-Ashkar D. The prevalence of depressive symptoms and its socioeconomic determinants among university students in Al Ain, UAE. Int J Pharm Pharm Sci 2014;6(5):4.
- 10. Razzak HA, Harbi A, Ahli S. Depression: prevalence and associated risk factors in the United Arab Emirates. Oman Med J 2019 Jul;34(4):274-282.
- 11. Lesch KP, Bengel D, Heils A, Sabol SZ, Greenberg BD, Petri S, et al. Association of anxiety-related traits with a polymorphism in the serotonin transporter gene regulatory region. Science 1996 Nov;274(5292):1527-1531.
- 12. Sen S, Burmeister M, Ghosh D. Meta-analysis of the association between a serotonin transporter promoter polymorphism (5-HTTLPR) and anxiety-related personality traits. Am J Med Genet B Neuropsychiatr Genet 2004 May;127B(1):85-89.
- 13. Hettema JM, Webb BT, Guo A-Y, Zhao Z, Maher BS, Chen X, et al. Prioritization and association analysis of murine-derived candidate genes in anxiety-spectrum disorders. Biol Psychiatry 2011 Nov;70(9):888-896.
- 14. Otowa T, Hek K, Lee M, Byrne EM, Mirza SS, Nivard MG, et al. Meta-analysis of genome-wide association studies of anxiety disorders. Mol Psychiatry 2016 Oct;21(10):1391-1399.
- 15. Dannlowski U, Ohrmann P, Konrad C, Domschke K, Bauer J, Kugel H, et al. Reduced amygdala-prefrontal coupling in major depression: association with MAOA genotype and illness severity. Int J Neuropsychopharmacol 2009 Feb;12(1):11-22.
- 16. Schulze TG, Müller DJ, Krauss H, Scherk H, Ohlraun S, Syagailo YV, et al. Association between a functional polymorphism in the monoamine oxidase A gene promoter and major depressive disorder. Am J Med Genet 2000 Dec;96(6):801-803.
- 17. Yu YW, Tsai S-J, Hong C-J, Chen T-J, Chen M-C, Yang C-W. Association study of a monoamine oxidase a gene promoter polymorphism with major depressive disorder and antidepressant response. Neuropsychopharmacology 2005 Sep;30(9):1719-1723.
- 18. Devang N, Satyamoorthy K, Rai PS, Nandini M, Rao S, Phani NM, et al. Association of HSD11B1 gene polymorphisms with type 2 diabetes and metabolic syndrome in South Indian population. Diabetes Res Clin Pract 2017 Sep;131:142-148.
- 19. Devang N, Satyamoorthy K, Rai PS, Nandini M, Basu A, Adhikari P. Association of HSD11B1 rs12086634 and HSD11B1 rs846910 gene polymorphisms with polycystic ovary syndrome in South Indian women. Int J Diabetes Dev Ctries 2018;38(4):381-386.
- 20. West J. An Arabic validation of a depression inventory. Int J Soc Psychiatry 1985;31(4):282-289.
- 21. AlHadi AN, AlAteeq DA, Al-Sharif E, Bawazeer HM, Alanazi H, AlShomrani AT, et al. An arabic translation, reliability, and validation of patient health questionnaire in a Saudi sample. Ann Gen Psychiatry 2017 Sep;16(1):32.
- 22. Kroenke K, Spitzer RL, Williams JB. The PHQ-9: validity of a brief depression severity measure. J Gen Intern Med 2001 Sep;16(9):606-613.
- 23. Beck AT, Steer RA, Brown GK. Manual for beck depression inventory-II. San Antonio, TX: Psychological Corporation; 1996.
- 24. Titov N, Dear BF, McMillan D, Anderson T, Zou J, Sunderland M. Psychometric comparison of the PHQ-9 and BDI-II for measuring response during treatment of depression. Cogn Behav Ther 2011;40(2):126-136.
- 25. Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998 Mar;92(6):829-839.
- 26. Scarpulla RC. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene 2002 Mar;286(1):81-89.
- 27. St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jäger S, et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 2006 Oct;127(2):397-408.
- 28. Tsunemi T, Ashe TD, Morrison BE, Soriano KR, Au J, Roque RA, et al. PGC-1α rescues Huntington’s disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci Transl Med 2012 Jul;4(142):142ra97.
- 29. Sweeney G, Song J. The association between PGC-1α and Alzheimer’s disease. Anat Cell Biol 2016 Mar;49(1):1-6.
- 30. Frenken H. Neuroticism, stress and psychological wellbeing: the role of CRHR1 and HSD11B1 in neuroticism, anxiety and depression. Edinburgh Research Archive 2012.