| |
Abstract
Objective: Chronic obstructive pulmonary disease (COPD) is a major health problem. The disease is driven by abnormal inflammatory reactions in response to inhaled particles and fumes. Therefore, inflammatory mediators are postulated to be of distinct importance. Keeping in view of the above facts; we investigate the role of polymorphisms of cytokine genes in the genetic predisposition of COPD.
Methods: In this present case-control study, the allele and genotype distributions of IL1B, IL1RN, TNF-α, and IL4 were studied in COPD patients (N=204) and healthy individuals (N=208). Genomic DNA was obtained by whole blood and genotyping was carried out by a polymerase chain reaction (PCR) based Restriction Fragment Length Polymorphism technique.
Results: Genotype IL1RN*2/IL1RN*2 was identified as protective for male COPD, its frequency being 8.7% in COPD patients and 14.6% in healthy subjects (p=0.017; OR=0.53), but IL1RN*1/IL1RN*2 turned out to be a risk factor for females COPD. No significant differences were found between the groups of COPD patients and healthy subjects concerning the genotype frequencies of the polymorphisms T (-511) C of IL1B and 70bp VNTR of IL-4. Genotype GA of the TNF-α polymorphism G (-308) A was more common in the COPD patients than in the controls (20.5% vs.14.4%; p=0.107), and allele A was significantly associated with COPD patients (p=0.023; OR=0.65).
Conclusion: IL-1RN *2 allele appears to be significantly associated with the COPD female patients and TNF-α-308A allele is a risk factor for the development of COPD.
Keywords: Chronic obstructive pulmonary disease (COPD); Interleukine-1 (IL-1); Tumor necrosis factor - alpha (TNF- 308).
Introduction
Chronic obstructive pulmonary disease (COPD), includes chronic pulmonary emphysema (CPE) and chronic bronchitis.1 Cigarette smoking is the known risk factor, ~ 15% of cigarette smokers developed COPD.2 Tobacco smoke activates cells of the respiratory pathway, thus triggering the whole inflammatory cascade.3,4 Cytokines are immunoregulatory substances that are synthesized by macrophages and lymphocytes.5 The polymorphism of the interleukin-1 (IL-1) gene cluster is implicated in the regulation of IL-1 and IL-1 receptor antagonist (IL-1RA) production,5 and is associated with several diseases linked to inflammation.6 Interleukin-1 receptor antagonist (IL1RN) binds to the IL1 receptor and acts as a competitive inhibitor of IL1b. It has also been revealed that a level of IL1b is increased in bronchoalveolar lavage fluid from chronic smokers in a cigarette dose-dependent manner.7 In mice, TNF-α overproduction has led to pulmonary emphysema and inflammation,8 and is thought to drive 70% of cigarette smoke-induced emphysema and inflammation.9 In COPD patients, there is a higher concentration of TNF-α in bronchial biopsies.10 A recent meta-analysis found an association between COPD and elevated serum TNF-α levels.11 Sakao et al. found the polymorphism TNF-α (-308) to be significantly associated with COPD in smokers,11,12 data from other studies have been conflicting.13,14
IL-4 is a prototypic member of Th2 cytokines and is a potent anti-inflammatory. It reduces the production of proinflammatory cytokines and destructive enzymes by monocytes.15 Previous studies reported that IL-4 inhibits the release of inflammatory mediators, such as TNF-α, IL-6, and IL-1α from activated monocytes.16 IL-4 VNTR gene polymorphism was conducted on COPD patients from two populations - Japanese and Egyptians but no significant association was observed.17
These contradictory results might be partly due to ethnic differences under different environmental conditions. Since the susceptibility to COPD is considered to be influenced by multiple genetic causes and gene-environment interactions,18 it is also possible that different polymorphisms in different ethnic groups cause the same COPD phenotype. However, it is still important to confirm the associations of the polymorphisms in different populations. So far, the relationship between genetic polymorphisms of cytokines and COPD has been controversial. In the present study, a case control association study was conducted to examine the role of polymorphisms of TNF-α, IL1ß, IL1RN and IL-4 genes and the development of COPD in a Northern Indian population.
Methods
All the subjects were the same as those in a previous study.19 A total of 204 COPD patients and 208 healthy controls were enrolled from the Outpatient Department of Pulmonary Medicine, Chhatrapati Shahuji Maharaj Medical University, Lucknow, India, after evaluation of lung function by spirometry. All the patients were diagnosed according to the Global Initiative for chronic obstructive lung disease (GOLD).20 The COPD group consisted of 179 males and 25 females (mean age: 57.83±10.58 years) and the healthy control group had 166 males and 42 females (mean age: 56.35±8.12 years). In the exclusion criteria; interstitial lung disease, malignancy, previous history of asthma, or tuberculosis were not considered. Healthy age-matched subjects without previous or present medical history of respiratory disorders or symptoms (persistent cough, sputum production, or dyspnea), seen by their practitioner for regular checkup, were recruited consecutively as controls. The participants were residents in the geographical area of northern India. The study was approved by the institutional ethical committee (Approval Code No-XXVIII ECM-B/P3), and the subjects were enrolled in accordance with the ethical standards of the Helsinki Declaration of 1975. All subjects gave their written informed consent.
Sample collection was done by taking venous blood sample (5 mL) from each patient and kept frozen until DNA extraction. The genomic DNA was extracted from peripheral blood leukocytes using the salting-out method.21
Genotyping for IL-1ß (-511 C>T) polymorphism was conducted using Polymerase chain reaction (PCR) in a total volume of 50 µl with 25 pmol of each primer; as reported by Rad et al.22 5 ‘TGGCATTGATCTGGTTCATC’ 3 and 5 ‘GTT TAGGAATCTTCCCACTT’ 3; genomic DNA (100-150 ng); 10 × Taq polymerase buffer (5 µl), and 1.5 units of Taq DNA polymerase (Bangalore Genei, India). PCR conditions were as follows: initial denaturation of 95°C for 5 minutes, 35 cycles of 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds, followed by a final extension of 72°C for 5 minutes. PCR products were digested by restriction endonuclease AvaI (MBI Fermentas) at 37°C overnight and then analyzed by 10% polyacrylamide gel electrophoresis using a 50-bp ladder. IL-1ß genotypes were coded as follows: 304 bp for T/T; 190 and 114 bp for C/C; 304+ 190 and 114 bp for C/T genotype.
Genotyping for IL-1RN-86bp (VNTR) polymorphism was done through PCR analysis which was carried out in a total volume of 50 µl containing genomic DNA (100-150 ng); 10 pmol of each primer; 10 × Taq polymerase buffers (5 µl), and 1.5 units of Taq DNA polymerase (Bangalore Genei). The primers for the IL-1RN 86bp (VNTR) in intron 2 were 5‘CTCAGCAACACTCCTAT’ 3 and 5 ‘TCCTGGTCTGCAGGTAA’ 3, according to Tarlow et al.23 Amplification was performed under the following conditions: denaturing step at 95°C for 5 minutes, 35 cycles of 95°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds and 1 cycle of extension at 72°C for 10 minutes. The PCR products were separated by electrophoresis on a 2% agarose gel. Sizing of product was performed using a 50-bp ladder. Allele 1 (4 repeats) was 410 bp, allele 2 (2 repeats) 240 bp, allele 3 (3 repeats) 325 bp, allele 4 (5 repeats) 500 bp, and allele 5 (6 repeats) 595 bp.
While genotyping for TNF-α(-308 G>A) polymorphism was done by ARMS-PCR using specific primers as described.24 Amplified products were checked on 10% PAGE with molecular weight markers. Band size of 224 bp showed the presence of G allele while 154 bp represented A allele. A control band of 323 bp was also observed in all PCR products.
The genotyping for IL-4 -70bp (VNTR) polymorphism was also conducted. The IL4 variable number of tandem repeat (VNTR) has been described as a 70-bp repeat in intron 3.25 The primers used were 5-TAGGCTGAAAGGGGGAAAGC-3 and 5-CTGTTCACCTCAACTGCTCC-3. The amplification consisted of 27 cycles of 0.5 min at 95°C, 0.7 min at 55°C and 0.7 min at 72°C. Allelic fragments were resolved on either 3% agarose gels electrophoresis. Alleles were named as follows: allele 1=three repeats (254 bp), and allele 2=two repeats (184 bp).
Descriptive statistics of patients and controls were presented as mean and standard deviations for continuous measures whereas frequencies and percentages were used for categorical measures. Effective sample sizes were calculated by the Quanto software version 1.2 (Gauderman WJ, Morrison JM.QUANTO 1.1 a computer program for power and sample size calculations for genetic-epidemiology studies (http://hydra.usc.edu/gxe) (2006). Independent sample t-test was performed to compare the mean values of continuous data and χ2-test was used to compare the genotype frequencies between patient and control. The clinical parameters were expressed as Mean ± SD. Odds ratio (OR), 95% confidence interval (CI) and p-value of genotype data were adjusted for confounding factors such as age, sex and pack-years of smoking. A p value of £0.05 was considered statistically significant, Hardy-Weinberg equilibrium using the goodness-of-fit χ2 test. Statistical analysis of the IL-1ß and IL-1RN haplotype frequency and linkage disequilibrium calculations were conducted using Arlequin software ver. 2.00 by the expectation-maximization algorithm.26 Genotype and haplotype risk was expressed as an age- and gender-adjusted OR (odds ratio) with 95% confidence interval (CI) estimated using binary logistic regression statistics. Bonferroni correction (a multiple-comparison correction) was applied to significant associations (p [pcorr]) for avoiding the type 1 error. The collected data were analyzed using the statistical software SPSS, ver. 15.0 (SPSS, Inc., Chicago, IL, USA).
Results
The characteristics of all participants in this study are shown in Table 1. Patients having FEV1 >70% were excluded from the COPD group in this study. All the patients in this study have all the stages of COPD according to GOLD classification of severity.27 In the control group, all pulmonary function parameters were within the normal range. No significant differences were observed in age. Significant differences were found in pack-year smoked (p=0.001); in the controls, it was 13.51±6.97 pack-years, but patients with COPD had a 23.67±17.89 pack-year history of smoking. In COPD patients, significant lower lung function was observed as compared with healthy controls, their pulmonary function tests showed predicted mean post bronchodilator FEV1 of 29.24±12.96%, and predicted (FVC) of 53.31±12.90%, with mean FEV1/FVC ratio of 54.85±14.22%, while the healthy controls showed FEV1 of 80.82±23.36%, FVC 84.75±19.80%, with a mean of FEV1/FVC ratio 94.54±14.04%.
Table 1: Demographic characteristic of COPD cases and controls.
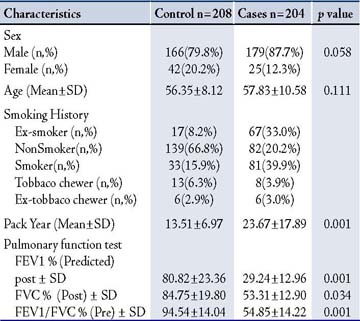
In terms of the distribution of IL-1ß (-511C>T) and IL-1RN 70bp (VNTR) Genotypes in COPD Patients and Controls; in the controls, the frequency of IL-1 ß (-511C>T) genotypes CC, CT, and TT was 11.1%, 48.6%, and 40.4% and the frequency of IL-1RN (86bp VNTR) genotypes 1/1, 1/2, 2/2, 1/3, and 1/5 was 51.0%, 35.6%, 11.5%, 1.4% and 0.5%, respectively. The observed frequency did not differ significantly from controls and followed the Hardy-Weinberg equilibrium (p=ns). Also, the distribution of IL-1ß (-511C>T) and IL-1RN (86bp VNTR) and IL-1ß (-511C>T) genotypes did not differ between COPD patients and controls. (Tables 2)
The genotypic frequency of the heterozygous genotype in TNF-α 308G>A polymorphism was higher in COPD patients (20.5%) as compared to the healthy controls (14.4%) but showed no significant association (p=0.107; OR=0.65; 95% CI=0.39-1.09) to the genotypes but frequency of A allelic was higher in COPD cases as compared with the healthy controls (10.2%) vs. (7.2%), and showed significant association (OR; 0.65, 95% CI=0.45-0.94, p=0.023). The distribution of IL-4 (VNTR) genotype in COPD patients exhibited decreased frequency of 1/1 genotypes (3.9% in patients vs. 6.3% in controls), slightly increased frequency of 1/2 genotypes (33.3% in patients vs. 28.4% in controls) but the differences were not significant (OR=1.93; 95% CI=0.95-3.93; p=0.071).
Table 2: Distribution of genotype and allele frequencies of IL-1β -511C>T, TNF-α 308 G>A, IL-1RN (86bp VNTR) and IL-4 (70bp VNTR) by the case-control status and association with risk of COPD Patients.
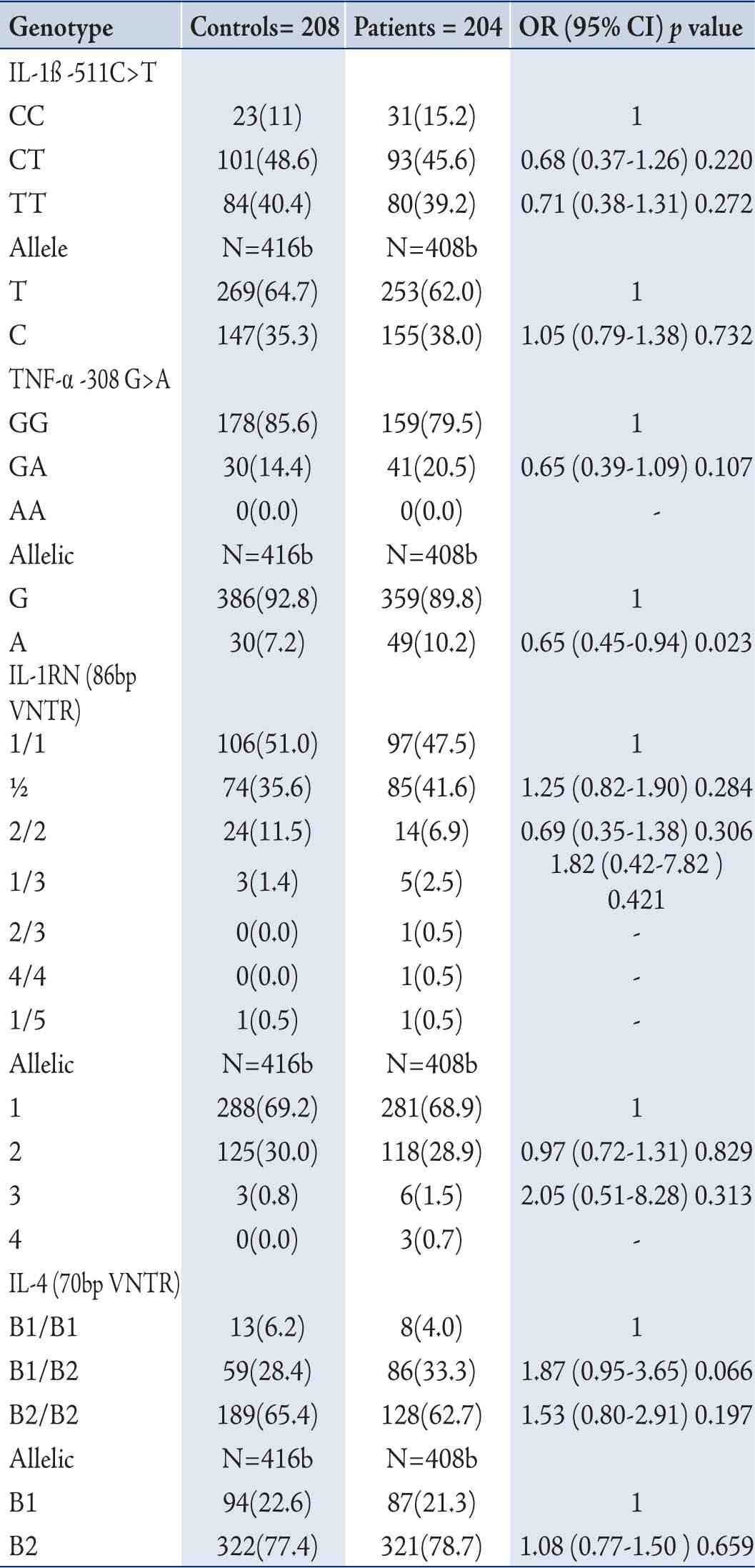
Linkage disequilibrium existed between IL-1ß (-511C>T) and IL-1RN (86 bp VNTR) polymorphisms in patient (χ2=4.76, p=0.064), and the controls (χ2=10.79, p=0.139). In haplotype analysis, six haplotypes, C1, C2, C3, T1, T2, and T3, were observed from the genotype combination of IL-1ß (-511C>T) and IL1-RN (86bp VNTR) polymorphisms. None of the haplotype combinations demonstrated significant risk of COPD after adjusting for age and gender. (Table 3)
Table 3: Distribution of IL-1ß (-511 C>T) and IL1RN (86bp VNTR) haplotype and risk of COPD.
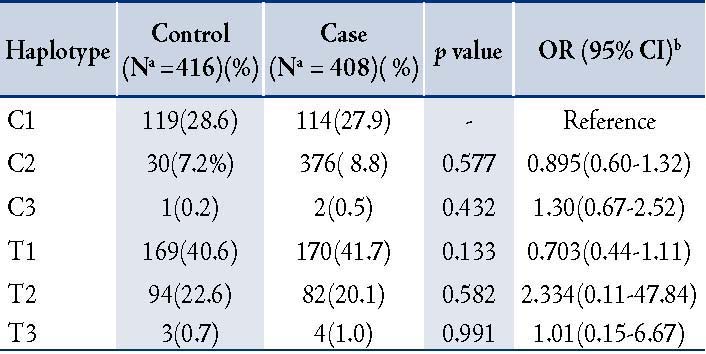
Table 4: Distribution of genotype and allele frequencies of IL-1β -511C>T and IL-1RN (86bp VNTR) in COPD patients and controls (Male and Female)
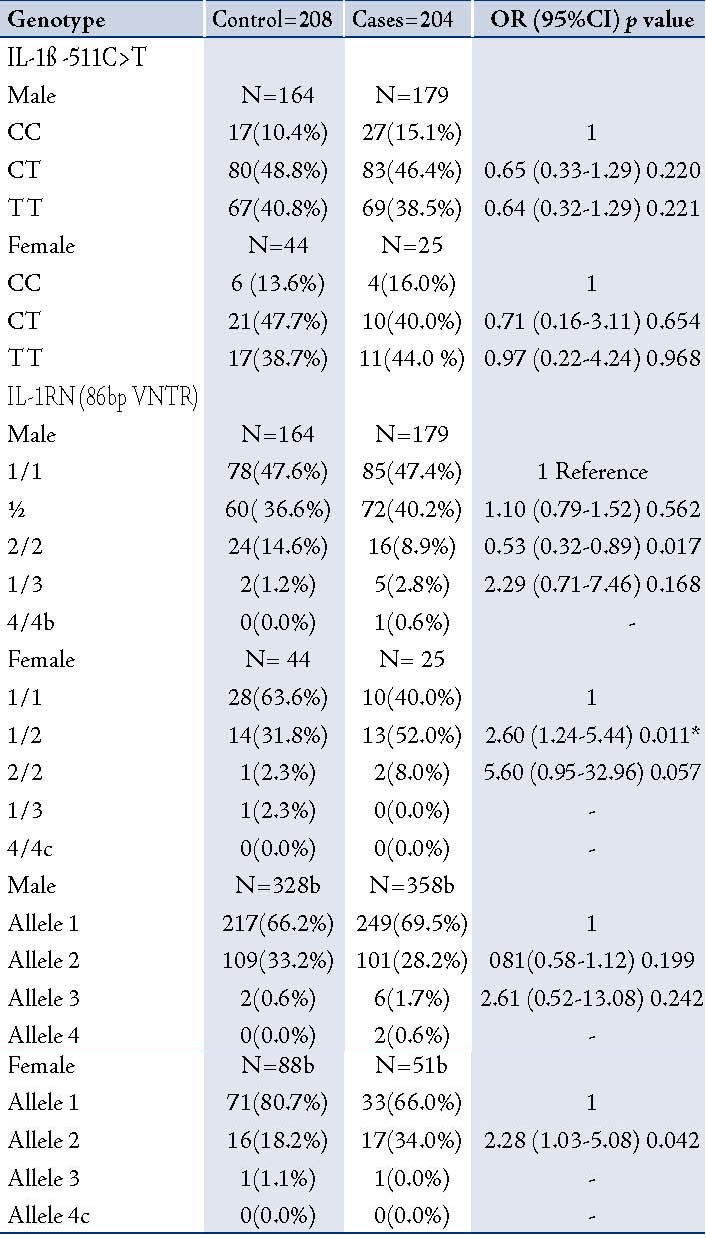
In order to determine the distribution of IL-1ß (-511C>T) and IL-1RN 86bp (VNTR), TNF-α (-308G>A), and IL-4 (70bp VNTR) genotypes in male and female COPD patients and healthy controls; the study participants were stratified according to gender and genotypic/allele frequencies of male and female COPD patients were compared with male and female healthy controls. There was no significant difference in polymorphism in -511C>T as compared to healthy male and female healthy controls. In IL1RN (86bp VNTR) polymorphism after gender stratification, a significant association was found with the female COPD patients as compared with healthy female controls. This association was strong and persisted even after the Bonferroni correction, (pcorr=0.022), (OR=5.60; 0.95-32.96; p=0.057), (Table 4). The interaction of male-female subjects with other polymorphisms did not show significant differences between cases and controls (data not shown).
Gene-environment interaction was detected on analyzing the interaction of genotypes with smoking habits, smokers with the IL4 (70bp VNTR) B1/B2 genotype where the risk for developing COPD was increased by more than two-fold (OR=5.12; 95%CI=1.14-24.04; p=0.033). However, after applying Bonferroni correction, the significance disappeared (Data not shown). The interaction of other polymorphisms with smoking habits showed no modulation of risk for developing COPD.
Discussion
This study investigated whether TNF-α, IL1ß, IL1RN, and IL-4 gene polymorphisms are risk factors for the development of COPD. No significant differences were found for IL1ß -511 C/T and IL4 (70bp VNTR) polymorphisms between COPD patients and healthy controls. In TNF-α (-308G>A), a significant difference was observed at allele level between the COPD patients and healthy controls. For IL-1RN VNTR although no overall association was observed, but after stratification based on gender, subgroup analysis genotype ½ and allele 2 were significantly associated with female COPD patients compared to female healthy controls.
The frequency distribution of IL-1ß (-511C/T) genotypes in the control population was similar to Japanese and Caucasian populations and a northern Indian population.28 In previous studies conducted in Japan, United States and Netherlands, no association was seen with the IL1ß C(-511)T and IL1RN (86 bp VNTR) polymorphisms.29,30
In the current study, IL1RN* 2 allele frequency in female COPD subjects was higher than in healthy female controls (OR, 2.28; 95% CI, 1.03-5.08; p=0.042). The exact reason for sex-based risk is not understood. Van der Strate et al. found that autoimmune component may be present in the development of COPD,31 8% of the population is affected by autoimmune diseases, 78% of whom are women. Women are known to respond to infection, vaccination, and trauma with increased antibody production and a T helper (Th) 2-predominant immune response, whereas a Th1 response is predominant in men. The onset of menopause also alters the immune system in healthy females,32 yet the impact of this alteration on lung function in women who smoke is unknown. Other mechanisms underlying differential disease susceptibility may include sex differences in nicotine metabolism and other cigarette by-products, and the influence of sex hormones on the biological response to risk factors.33 It is known that allele IL1RN*2 is associated with higher plasma levels of IL1RN as compared with IL1RN*1,34 and IL1RN*2 allele is also associated with increased production of IL1RN,35 and IL1b.36 The production of IL-1RA, as well as IL-1, appears to differ between men and women,37 in vitro production of IL-1RA from isolated monocytes was significantly higher in women than in men.38 It may however, be added that the total number of female COPD with IL1RN* 2 were limited in the present study. Therefore, it may be considered as a trend but replicated in other population with larger sample size.
In this study, TNF-α 308A allele frequencies were 10.2% and 7.2% in the patients and the controls, and significantly associated (OR, 0.65; 95% CI, 0.45-0.94; p=0.023) with COPD. There are several case-control studies investigating the relationship of TNF-α gene polymorphisms and the development of COPD,39 but most of them9,16 demonstrated no association of TNF-α -308 G/A with COPD. The frequencies of the TNF-α 308 G/A polymorphism in this study were consistent with the previous data reported by others.40 Zhan et al. conducted a meta-analysis, and found that the TNF-α -308 polymorphism was significantly associated with an increased risk of COPD, and the A allele appeared to be a more significant risk factor for developing COPD among Asians but not in Caucasian populations.12,39 Thus, this study again confirms TNF-308 A to be a risk factor for COPD in Asians.
IL-4 is a prototypic member of Th2 cytokines and is a potent anti-inflammatory. It reduces the production of proinflammatory cytokines and destructive enzymes by monocytes.15 The IL-4 gene is located on the long arm of chromosome 5 (q23–31) together with other Th2 cytokine genes and is present in a cluster of cytokine genes (IL-3, -5, -9, -13, and -15, granulocyte colony-stimulating factor, and interferon regulatory factor).41 A gene contains a variable number of tandem repeat polymorphism located in the third intron.25 Similar to this study findings, the VNTR polymorphism has not been found to be associated with COPD in various other studies.42
Conclusion
The present study illustrated that IL-1ß (-511C>T) and IL-4 (70 bp VNTR) both at the allele or genotype level had no influence on the overall susceptibility of COPD patients. The TNF-α 308 A allele was also significantly associated with the COPD patients. However, a sex specific difference in the IL-1RN *2 allele appear to be significantly associated with the COPD female patients. IL-1RN*2 allele may represent a potential tool to assess the gender specific susceptibility in COPD patients. However, COPD is a mutifactorial disease with very high environmental contribution. Therefore, genetic analysis of other cytokine genes and their interactions among themselves and environmental factors may also be required to better understand this complex interplay of gene polymorphisms in the development of COPD.
In this study, sample size was one of the majors limiting factors. In future, replication studies with larger sample size in different populations are desired for definitive associations and future applications.
Acknowledgements
The study was supported by a research grant from Council of Science & Technology U.P., Lucknow (UPCST/SERPD-D-3404) and Indian Council of Medical Research, New Delhi.
References
1. American Thoracic Society. Standards for the diagnosis and care of patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med 1995 Nov;152(5 Pt 2):S77-S121.
2. Bascom R. Differential susceptibility to tobacco smoke: possible mechanisms. Pharmacogenetics 1991 Nov;1(2):102-106.
3. Oudijk EJ, Nijhuis EH, Zwank MD, van de Graaf EA, Mager HJ, Coffer PJ, et al. Systemic inflammation in COPD visualised by gene profiling in peripheral blood neutrophils. Thorax 2005 Jul;60(7):538-544.
4. Barnes PJ, Shapiro SD, Pauwels RA. Chronic obstructive pulmonary disease: molecular and cellular mechanisms. Eur Respir J 2003 Oct;22(4):672-688.
5. Vamvakopoulos J, Green C, Metcalfe S. Genetic control of IL-1beta bioactivity through differential regulation of the IL-1 receptor antagonist. Eur J Immunol 2002 Oct;32(10):2988-2996.
6. Witkin SS, Gerber S, Ledger WJ. Influence of interleukin-1 receptor antagonist gene polymorphism on disease. Clin Infect Dis 2002 Jan;34(2):204-209.
7. Kuschner WG, D’Alessandro A, Wong H, Blanc PD. Dose-dependent cigarette smoking-related inflammatory responses in healthy adults. Eur Respir J 1996 Oct;9(10):1989-1994.
8. Lundblad LK, Thompson-Figueroa J, Leclair T, Sullivan MJ, Poynter ME, Irvin CG, et al. Tumor necrosis factor-alpha overexpression in lung disease: a single cause behind a complex phenotype. Am J Respir Crit Care Med 2005 Jun;171(12):1363-1370.
9. Churg A, Wang RD, Tai H, Wang X, Xie C, Wright JL. Tumor necrosis factor-alpha drives 70% of cigarette smoke-induced emphysema in the mouse. Am J Respir Crit Care Med 2004 Sep;170(5):492-498.
10. Mueller R, Chanez P, Campbell AM, Bousquet J, Heusser C, Bullock GR. Different cytokine patterns in bronchial biopsies in asthma and chronic bronchitis. Respir Med 1996 Feb;90(2):79-85.
11. Gan WQ, Man SF, Senthilselvan A, Sin DD. Association between chronic obstructive pulmonary disease and systemic inflammation: a systematic review and a meta-analysis. Thorax 2004 Jul;59(7):574-580.
12. Sakao S, Tatsumi K, Igari H, Shino Y, Shirasawa H, Kuriyama T. Association of tumor necrosis factor alpha gene promoter polymorphism with the presence of chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2001 Feb;163(2):420-422.
13. Chierakul N, Wongwisutikul P, Vejbaesya S, Chotvilaiwan K. Tumor necrosis factor-alpha gene promoter polymorphism is not associated with smoking-related COPD in Thailand. Respirology 2005 Jan;10(1):36-39.
14. Seifart C, Dempfle A, Plagens A, Seifart U, Clostermann U, Müller B, et al. TNF-alpha-, TNF-beta-, IL-6-, and IL-10-promoter polymorphisms in patients with chronic obstructive pulmonary disease. Tissue Antigens 2005 Jan;65(1):93-100.
15. Chomarat P, Vannier E, Dechanet J, Rissoan MC, Banchereau J, Dinarello CA, et al. Balance of IL-1 receptor antagonist/IL-1 beta in rheumatoid synovium and its regulation by IL-4 and IL-10. J Immunol 1995 Feb;154(3):1432-1439.
16. Lee H, Clark B, Gooi HC, Stoves J, Newstead CG. Influence of recipient and donor IL-1alpha, IL-4, and TNFalpha genotypes on the incidence of acute renal allograft rejection. J Clin Pathol 2004 Jan;57(1):101-103.
17. Hegab AE, Sakamoto T, Saitoh W, Nomura A, Ishii Y, Morishima Y, et al. Polymorphisms of TNFalpha, IL1beta, and IL1RN genes in chronic obstructive pulmonary disease. Biochem Biophys Res Commun 2005 Apr;329(4):1246-1252.
18. Lander ES, Schork NJ. Genetic dissection of complex traits. Science 1994 Sep;265(5181):2037-2048.
19. Shukla RK, Kant S, Bhattacharya S, Mittal B. Association of genetic polymorphism of GSTT1, GSTM1 and GSTM3 in COPD patients in a north Indian population. COPD 2011 Jun;8(3):167-172.
20. Global Initiative for Chronic Obstructive Lung Disease (GOLD). Global strategy for the diagnosis, Management and prevention of chronic obstructive pulmonary disease. Updated 2009. http://www.goldcopd.com/ Accessed October 2010
21. Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988 Feb;16(3):1215.
22. Rad R, Prinz C, Neu B, Neuhofer M, Zeitner M, Voland P, et al. Synergistic effect of Helicobacter pylori virulence factors and interleukin-1 polymorphisms for the development of severe histological changes in the gastric mucosa. J Infect Dis 2003 Jul;188(2):272-281.
23. Tarlow JK, Blakemore AI, Lennard A, Solari R, Hughes HN, Steinkasserer A, et al. Polymorphism in human IL-1 receptor antagonist gene intron 2 is caused by variable numbers of an 86-bp tandem repeat. Hum Genet 1993 May;91(4):403-404.
24. Ye S, Dhillon S, Ke X, Collins AR, Day IN. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res 2001 Sep;29(17):E88-E8.
25. Mout R, Willemze R, Landegent JE. Repeat polymorphisms in the interleukin-4 gene (IL4). Nucleic Acids Res 1991 Jul;19(13):3763.
26. Schneider SR. Excoffier L Arlequin ver. 2.00. Software for population genetics data analysis. Switzerland: Genetics and Biometry Laboratory,University of Geneva 2000.
27. Global Initiative for Chronic Obstructive Lung Disease (GOLD). Global strategy for the diagnosis, Management and prevention of chronic obstructive pulmonary disease. Updated 2009. http://www.goldcopd.com/ Accessed October 2010.
28. Upadhyay R, Jain M, Kumar S, Ghoshal UC, Mittal B. Potential influence of interleukin-1 haplotype IL-1 beta-511*T-IL-1RN*1 in conferring low risk to middle third location of esophageal cancer: a case-control study. Hum Immunol 2008 Mar;69(3):179-186.
29. Joos L, McIntyre L, Ruan J, Connett JE, Anthonisen NR, Weir TD, et al. Association of IL-1beta and IL-1 receptor antagonist haplotypes with rate of decline in lung function in smokers. Thorax 2001 Nov;56(11):863-866.
30. Asada M, Yamaya M, Ebihara S, Yasuda H, Tomita N, Kubo H, et al. Interleukin-1beta gene polymorphisms associated with COPD. Chest 2005 Aug;128(2):1072-1073, author reply 1073.
31. van der Strate BW, Postma DS, Brandsma CA, Melgert BN, Luinge MA, Geerlings M, et al. Cigarette smoke-induced emphysema: A role for the B cell? Am J Respir Crit Care Med 2006 Apr;173(7):751-758.
32. Mund E, Christensson B, Larsson K, Grönneberg R. Sex dependent differences in physiological ageing in the immune system of lower airways in healthy non-smoking volunteers: study of lymphocyte subsets in bronchoalveolar lavage fluid and blood. Thorax 2001 Jun;56(6):450-455.
33. Camp PG, O’Donnell DE, Postma DS. Chronic obstructive pulmonary disease in men and women: myths and reality. Proc Am Thorac Soc 2009 Sep;6(6):535-538.
34. Hurme M, Santtila S. IL-1 receptor antagonist (IL-1Ra) plasma levels are co-ordinately regulated by both IL-1Ra and IL-1beta genes. Eur J Immunol 1998 Aug;28(8):2598-2602.
35. Danis VA, Millington M, Hyland VJ, Grennan D. Cytokine production by normal human monocytes: inter-subject variation and relationship to an IL-1 receptor antagonist (IL-1Ra) gene polymorphism. Clin Exp Immunol 1995 Feb;99(2):303-310.
36. Santtila S, Savinainen K, Hurme M. Presence of the IL-1RA allele 2 (IL1RN*2) is associated with enhanced IL-1beta production in vitro. Scand J Immunol 1998 Mar;47(3):195-198.
37. Lynch EA, D.C., Cannon JG Gender differences in IL-lao, IL-13, and IL-1 receptor antagonist secretion from mononuclear cells and urinary excretion. . J Immunol 1994 1994 153, 300-306
38. Kanai T, Fukuda-Miki M, Shimoya K, Azuma C, Hashimoto K, Nobunaga T, et al. Increased interleukin-1 and interleukin-1 receptor antagonist levels in cervical mucus in the ovulatory phase in comparison with the follicular phase. Gynecol Obstet Invest 1997;43(3):166-170.
39. Zhan P, Wang J, Wei SZ, Qian Q, Qiu LX, Yu LK, et al. TNF-308 gene polymorphism is associated with COPD risk among Asians: meta-analysis of data for 6,118 subjects. Mol Biol Rep 2011 Jan;38(1):219-227.
40. Ishii T, Matsuse T, Teramoto S, Matsui H, Miyao M, Hosoi T, et al. Neither IL-1beta, IL-1 receptor antagonist, nor TNF-alpha polymorphisms are associated with susceptibility to COPD. Respir Med 2000 Sep;94(9):847-851.
41. Le Beau MM, Lemons RS, Espinosa R III, Larson RA, Arai N, Rowley JD. Interleukin-4 and interleukin-5 map to human chromosome 5 in a region encoding growth factors and receptors and are deleted in myeloid leukemias with a del(5q). Blood 1989 Feb;73(3):647-650.
42. Nakashima H, Miyake K, Inoue Y, Shimizu S, Akahoshi M, Tanaka Y, et al. Association between IL-4 genotype and IL-4 production in the Japanese population. Genes Immun 2002 Apr;3(2):107-109.
|
|