| |
Abstract
Objective: Different findings indicate that CYP2C plays a clinical role in determining interindividual and interethnic differences in drug effectiveness. The ethnic differences in the frequency of CYP2C19 mutant alleles continue to be a significant study topic. The aim of the present study was to assess the frequency of allelic variants of CYP2C19 in Turkman ethnic groups and compare them with the frequencies in other ethnic populations.
Methods: The study group included 140 unrelated healthy ethnic Turkman subject referred to the Health Center. Genotyping of CYP2C19 alleles (CYP2C19*1, CYP2C19*2, and CYP2C19*3 alleles) was carried out by Polymerase Chain Reaction-Restriction Fragment Length Polymorphism technique
Results: The allele frequency of CYP2C19*1, CYP2C19*2 and CYP2C19*3 were 56.43%, 23.57% and 20%, respectively. The result also showed that 39.7% of subjects expressed the CYP2C19*1/*1 genotype. While 42.1%, 9.3%, 9.3% and 1.4% expressed CYP2C19*1/*2, CYP2C19*1/*3, CYP2C19*2/*2 and CYP2C19*3/*3 genotypes, respectively. The genotype CYP2C19*2/*3 was not expressed in this study population. The findings suggested that 10% of subjects were poor metabolizers by expressing CYP2C19*2/*2 and CYP2C19*3/*3 genotypes. Fifty one percent of subjects were intermediate metabolizers having CYP2C19*1/*2, CYP2C19*2/*3 and CYP2C19*1/*3 genotypes and 37.86% were found to be extensive metabolizers expressing CYP2C19*1/*1 genotype. The frequency of intermediate metabolizers genotype was high (51%) in Turkman ethnic groups.
Conclusion: This study showed that the determined allelic variants of CYP2C19 (CYP2C19*2 and CYP2C19*3 mutations) in Turkman ethnic group are comparable to other populations. These findings could be useful for the clinicians in different country to determine optimal dosage and effectiveness of drugs metabolized by this polymorphic enzyme.
Keywords: CYP2C19 genetic polymorphism; Iranian Turkman ethnic group; polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP).
Introduction
There are at least four isoforms of human CYP2C subfamily in mankind. The main forms are principally 2C8, 2C9, 2C18 and 2C19. Their related genes are located on chromosome 10.1,2 It has been shown that drug metabolism is directly related to genetic polymorphism and gene mutations manipulate the enzyme activities responsible for the drugs metabolism. Such enzyme activity modulation can present itself in three scenario of high, low and zero activities.3-5 From drug related genetic pattern, the human gene are sub-grouped into few phenotypes of poor (or slow) metabolizers (PM), intermediate metabolizers (IM), extensive (or rapid) metabolizers (EM), and ultrarapid metabolizers (UM).6 There are various reports of drugs which adversely affect human metabolism, resulting either from drug toxicity or long-lasting therapeutic effect of some drugs even consumption at therapeutic dosage in poor or slow metabolizer phenotypes. The ultra rapid metabolizer phenotype probably does not demonstrate the related therapeutic effect and this may be the reason why some drugs do not produce any therapeutic effect even in genetically susceptible subjects. This manifestation of specific phenotypes is related to the pharmocogeneticity of particular polymorphism of poor or slow, extensive or rapid and ultra-rapid metabolizer. It has been shown that about 3% of Caucasians express the poor metabolizer phenotypes S-mephenytoin, however literature review in this regards shows slight differences.7,8 Other studies indicated that East Asian subjects express the poor metabolizer phenotypes at higher frequency. Some researchers have indicated that 18-23% of Japanese,9,10 15-17% of Chinese11,12 and 12-16% Koreans express the poor metabolizer phenotype.13,14 The same index for Black African was reported to be 4-7%.15 Reports suggest that the poor metabolizer phenotype is an autosomal recessive trait which is inherited.8 Same reports have considered CYP2C19*2 and CYP2C19*3 to be the dominant poor metabolizer phenotype for malfunction of CYP2C19 alleles.16,17 Among East Asians, CYP2C19*2 is the major allele and it dominates about 75% of the defective alleles.16 These allele defections for Caucasians account for 93% of population.18 CYP2C19*3 is the phenotype which comprises approximately 25% of the defective gene among East Asians which was initially found in a Japanese poor metabolizer population.17 But it was discovered that the above mentioned phenotype is significantly rare among non-East Asian sub-population.19 The incidence of the poor metabolizer phenotype in European whites,7 and the residents of Vanuatu Island as well as Melanesia subjects,20 at a rate of 3-5% and 70%, respectively. The incidence of CYP2C19*2 and CYP2C19*3 alleles among north Indian subjects was reported to be as 29.7% and 0.00%, respectively.21 In an other study, the frequency of CYP2C19*2 and CYP2C19*3 alleles among South Indian of Tamil, Telgu, Kannada, and Malayalam backgrounds were reported to be 35% and 1%, respectively.22 The narrow therapeutic index of some drugs in particular are important safety in determining the drug metabolic capacity indications of some individual by applying the necessary phenotype and genotype standards to provide a safe guard for susceptible subjects. The aim of this study was to assess the distribution of CYP2C19 allele and genotypic variants among the Turkman ethnic group in comparison with other populations.
Methods
The study group included 140 unrelated healthy subjects of Turkman origin (people who speak Turkman as a native language and population inbreeding people) who were referred to the Health Center in Gonbadkavoos (located in North Eastern Iran, South East of the Caspian Sea). This present study, set to establish the prevalence of CYP2C19 variants in a sample of Turkman ethnic group and compare the collected data with other populations. Ethical approval was obtained from the ethics committee of Golestan University of Medical Sciences and informed consent was obtained from all participants.
Five milliliters of venous blood was sampled from each subject and collected into EDTA tubes. Extraction of DNA from peripheral white blood cells was done by salting out method.23 DNA extract was dissolved in sterilized distilled water and samples were kept in -20oC until analysis by polymerase chain reaction (PCR).
Genotyping of CYP2C19 alleles (CYP2C19*1, CYP2C19*2, and CYP2C19*3 alleles) was carried out by Polymerase Chain Reaction (PCR)-Restriction Fragment Length Polymorphism (RFLP) technique.24 PCR was done in 25 microliter mixture containing PCR buffer 10 mM Tris–HCl, pH 9, 1.5 mM MgCl2 (Fermentase, Burlington; Canada), 50 mM KCl (Fermentase, Burlington; Canada), 10 mM deoxyribonucleotide triphosphate (dNTP) mix, 5 U/µl Taq polymerase (Fermentase, Burlington; Canada), 5 pM of each primer (Bioneer; Korea), 500 ng DNA (Genomic; Korea) and sterile distillated water. PCR was carried out in a genetix CG palm-thermocycler (New Delhi; India). PCR products (10µl) were digested with restriction enzymes (Fermentase; Burlington; Canada), (SmaI for CYP2C19*2 and BamHI for CYP2C19*3) at 30oC and 37oC for 16 hrs for compelte digestion, respectively. Primers amplification was done as described by De Morais et al.17 The DNA fragments were electrophoresed (Apelex, France) on a 2% (for CYP2C19*3) and 3% (for CYP2C19*3) agarose gel and stained with Ethidium bromide. Bands were detected by a short wavelength UV transluminator, photographed using a Polaroid Gel Camera with Polaroid black and white film. The CYP2C19*2 mutation was detected using sense primer 5'-AATTACAACCAGAGCTTGGC-3' and antisense primer 5'-TATCACTTTCCATAAAAGCAAG-3'. The detection of CYP2C19*3 was carried out using sense primer 5'-AAATTGTTTCCAATCATTTAGCT-3' and antisense primer 5'-ACTTCAGGGCTTGGTCAATA-3'. The PCR amplification conditions for CYP2C9*2 and CYP2C9*3 were as follow: Initial denaturation, Number of cycle(s), Denaturation, Extention and Final extention step, which were 94oC, 300 sec.; 37; 94oC, 60 sec.; 72oC, 30 sec. and 72oC, 300 sec., respectively. The annealing temperature and time for CYP2C9*2 and CYP2C9*3 were 55oC, 30 sec. and 52oC, 45 sec., respectively. The Products of PCR before and after restriction enzyme digestion for CYP2C9*2 and CYP2C9*3 genotypes are summarized in Fig. 1 and 2, respectively.
Figure 1: PCR-restriction enzyme (SmaI digestion) fragmentation patterns on the agarose gel is stained by ethidium bromide for CYP2C19*2 from subjects representing *1/*1, *1/*2, *1/*2, and *1/*1 genotypes (From 2 to 5 wells, left to right on the agarose gel). DNA ladder was loaded in well 1 and the sizes of the PCR-restriction fragments were loaded in well 6.
Figure 2: PCR-restriction enzyme (BamHI digestion) fragmentation patterns on agarose gel is stained by ethidium bromide for CYP2C19*3 from subjects representing *1/*1, *1/*3, *1/*3, and *1/*1 genotypes (From 2 to 5 wells, left to right on the agarose gel). DNA ladder was loaded in well 1 and the sizes of the PCR-restriction fragments were loaded in well 6.
Statistical analysis was done to evaluate allele frequencies of CYP2C19 in Turkman subjects alongside other ethnic groups. The 95% confidence intervals (95% CI) for frequency of the variant alleles of each gene were determined. The observed genotype frequencies of CYP2C19 were observe and compared with expected frequencies according to the Hardy–Weinberg law. Differences in allele frequencies and PM genotype frequencies between the Turkman ethnic group and other populations from various area swere measured by Fisher exact test. Data was analysed using SPSS version 16.0. and statistical significance was considered at p<0.05.
Results
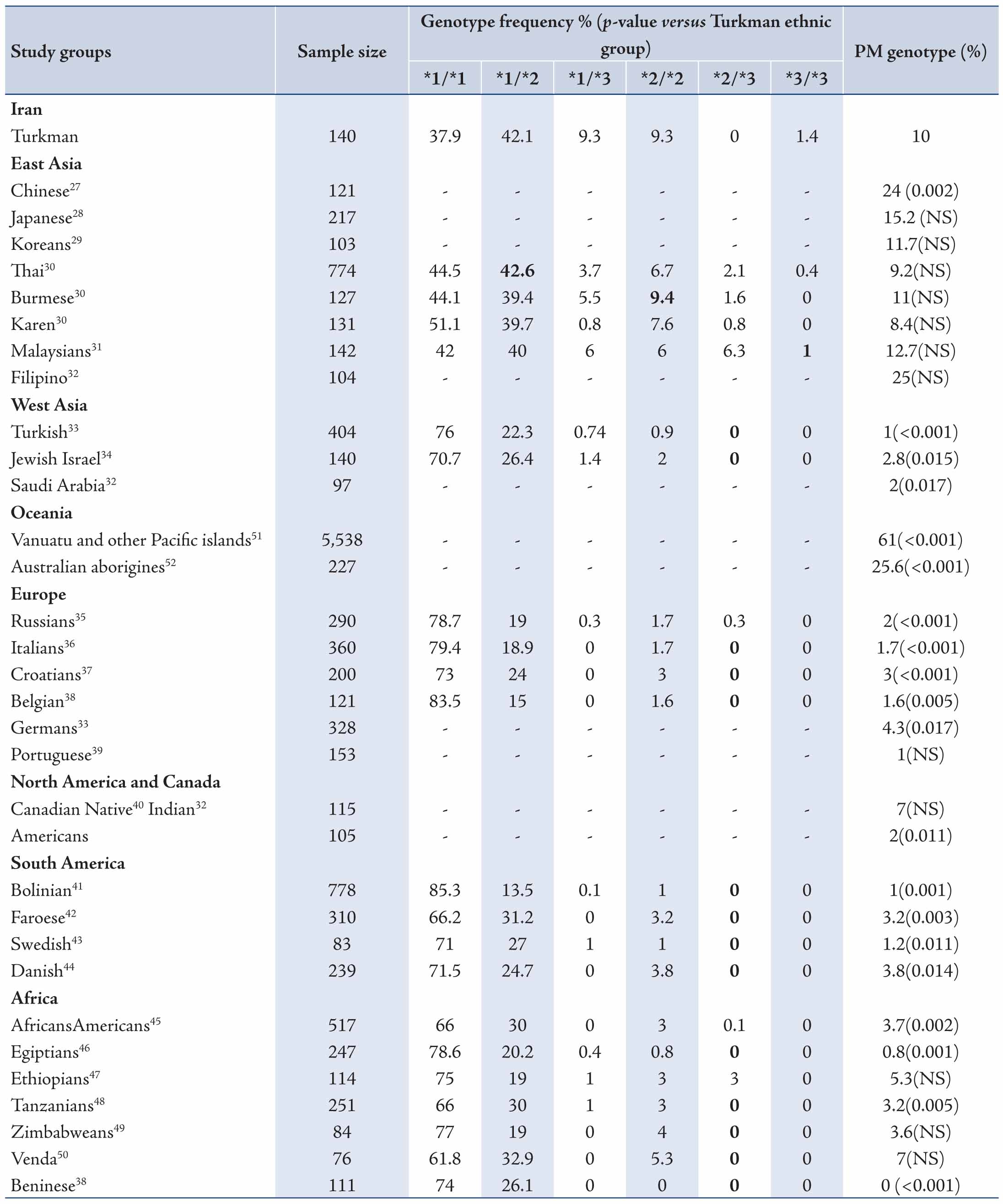
The data revealed mean ages of the participants to be 29.73±9.11 years old. Allele and genotype frequencies of CYP2C19 gene among the Turkman ethnic group are shown in Tables 1 and 2. The allele frequency of CYP2C19*1 (Wild type), CYP2C19*2 and CYP2C19*3 were 56.43% (95% CI: 48.15-64.36), 23.57% (95% CI: 17.31-31.25) and 20% (95% CI: 14.22-27.36), respectively. CYP2C19*1 was the most frequently (56.43%) determined allele in Turkman ethnic groups. The observed frequencies of CYP2C19 genotypes in Turkman ethnic group were found to be in the Hardy-Weinberg equilibrium (Table 2). The results showed that 39.7% of subjects expressed the CYP2C19*1/*1 genotype (95% CI: 30.25-46.11). While 59 (42.1%), 13 (9.3%), 13 (9.3%) and 2 (1.4%) subjects expressed the CYP2C19*1/*2 (95% CI: 34.28-50.42), CYP2C19*1/*3 (95% CI: 5.51-15.24), CYP2C19*2/*2 (95% CI: 5.51-15.24) and CYP2C19*3/*3 (95% CI: 0.3-5) genotypes, respectively. There was no expression CYP2C19*2/*3 (0%) genotype. Out of the alleles, CYP2C19*1/*2 (42.1%) was the most frequently observed mutant allele. Table 3 shows the prevalence of the predicted phenotypes of CYP2C19, which were as follow: 10.7% of subjects were PM carrying the CYP2C19*2/*2 and CYP2C19*3/*3 genotypes (95% CI: 5.5-15). Fifty one percent of subjects were IM carrying the CYP2C19*1/*2, CYP2C19*2/*3 and CYP2C19*1/*3 genotypes (95% CI: 43-60). Which 37.86% were found to be EM expressing the CYP2C19*1/*1 genotype (95% CI: 29.72-45.99). IM genotype frequency was high (51%) in the Turkman ethnic group.
Table 1: Allele frequencies of CYP2C19 gene among Turkman ethnic groups (n = 140).
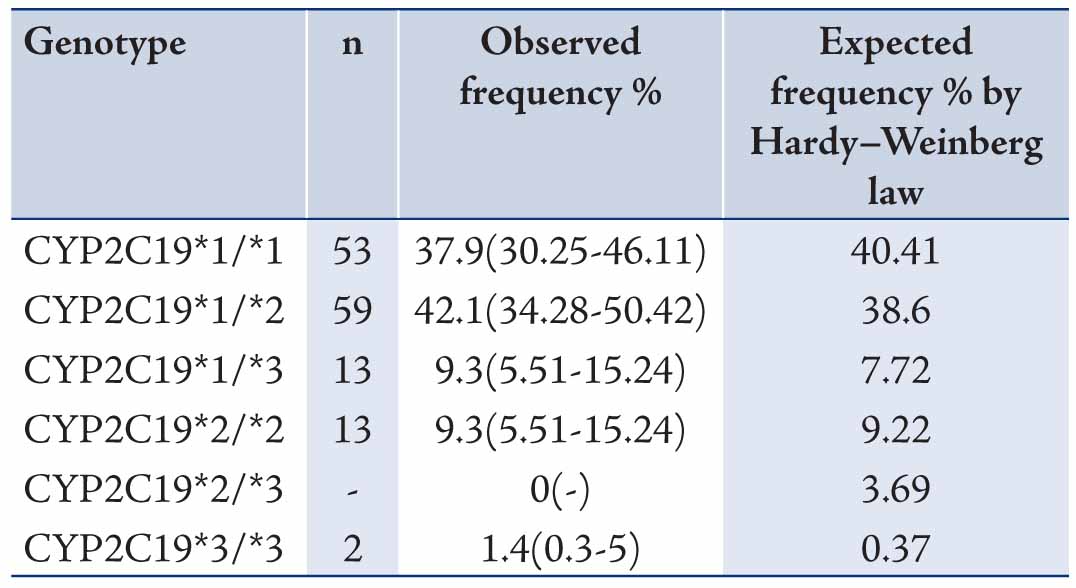
Table 2: Genotype frequencies of CYP2C19 gene among Turkman ethnic groups (n = 140).
Table 3: Prevalence of CYP2C19 predicted phenotypes in Turkman ethnic groups (n = 140) [χ2 test, p>0.05].
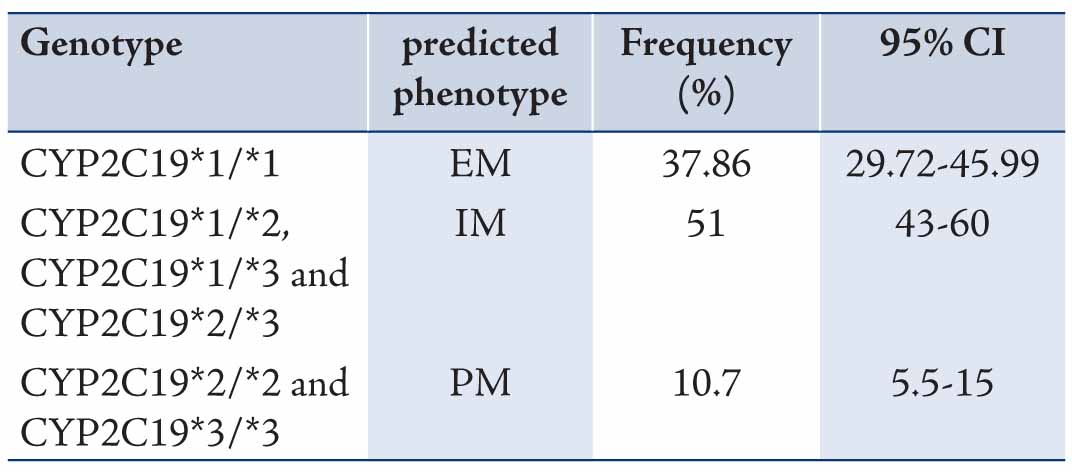
Table 4: Comparison of allele frequencies of CYP2C19 of the Turkman ethnic group with different populations.
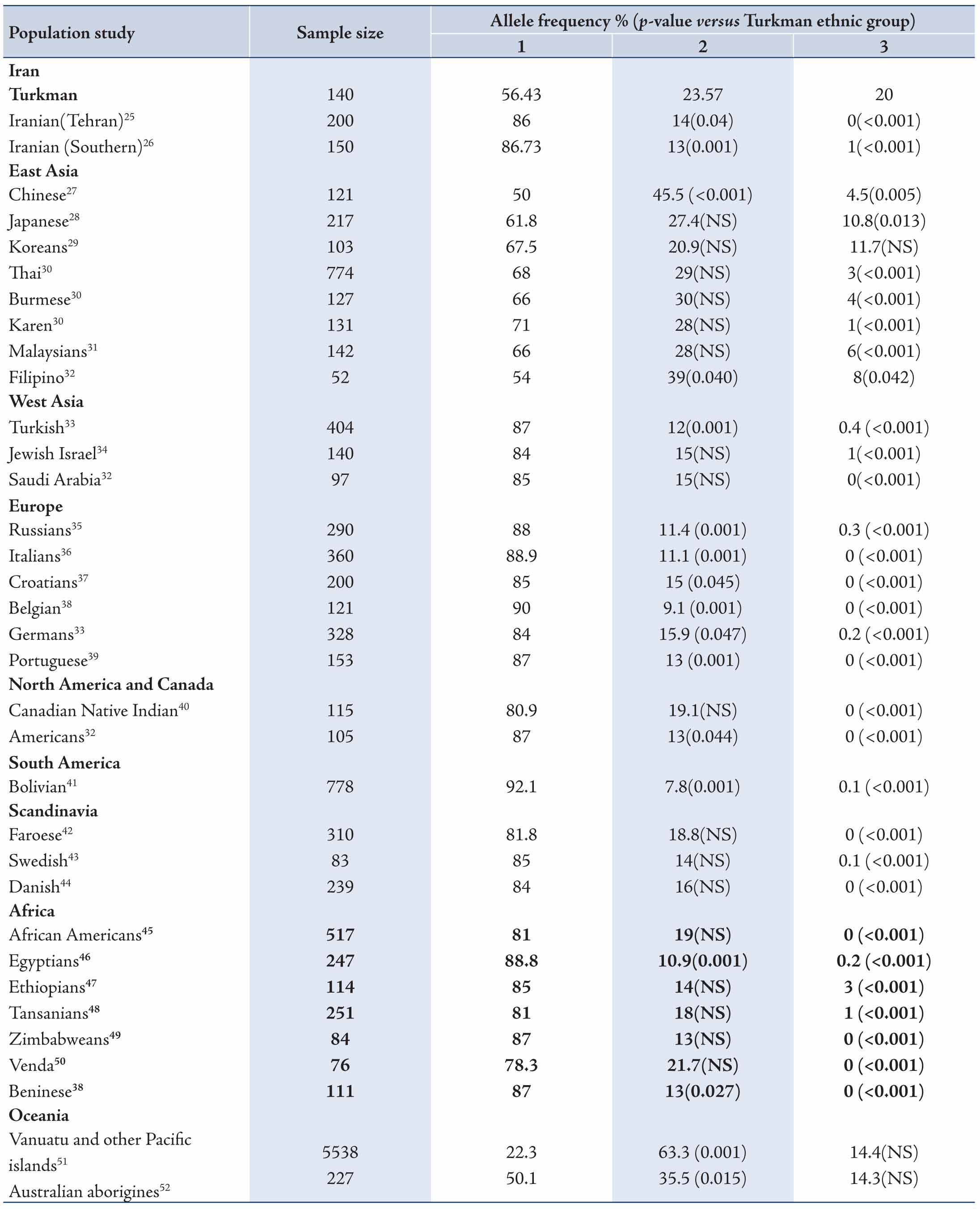
Table 5: Comparison of CYP2C19 genotype and poor metabolizer frequency between Turkman ethnic group and other populations.
Discussion
The Turkman ethnic group is a unique population in Iran, due to their population inbreeding, these people were considered to be particularly important in investigating the allele frequencies and genotype distributions of some variants of a pharmacogenetic interest. This is the first study based on the CYP2C19 genotype distribution in this ethnic group. There have been many reports on the genetic polymorphisms of CYP2C. Some studies have reported that the hydroxylation metabolism of S-mephenytoin showed genetic polymorphism.53,54 It was also determined that this enzyme catalyzes the metabolism of some drugs such as S-mephenytoin, methylphenobarbital, omeprazole, phenytoin, imipramine, proguanil, propranolol and diazepam.55 It has also reported that CYP2C shows a possible clinical role in determining interindividual and interethnic differences in drug effectiveness.
Change in CYP expression can affect drug response and activity. The ethnic differences in the frequency of CYP2C19 mutant alleles continues to be a significant study topic.56 In this current study, we assessed the distribution of CYP2C19 variants in the Turkman ethnic group and compared the data with those from other populations. The CYP2C19*2 variant was the most allele among Turkman ethnic groups. The frequency of this variant was 23.57% in the present study. Its frequency was lower than the figures reported from Chinese (45.5%),27 Japanese (27.4%),28 Thai (29%),30 Karen (28%),30 Malaysians (28%),31 Filipino (29%),32 Vanuatu and other Pacific islands (63.3%),51 as well as Australian Aborigines (35.5%).52 The frequency of CYP2C19*2 has been reported to range from 20.9% to 45.5%, 12%-15%, 35.5%-63.3%, 9.1%-15.9%, 13%-19.1%, 7.8%, 14%-18.8%, 10.9%-19% and 13%-14% in East Asian,27-32 West Asian,32-34 Oceanian,51,52 European,33,38,39 North American and Canadian,32,40 South American (Bolivian),41 Scandinavian,42-44 African38,45-50 and Iranian,25,26 respectively. The frequency of the CYP2C19*3 allele (20%) was high among the Turkman ethnic group when compared with some East and West Asians,27,31-34 Oceanian,51,52 European,33,38,39 North American and Canadian,32,40 South American (Bolivian),41 Scandinavian,42-44 African38,45-50 and Iranian25,26 populations, respectively. All combinations of alleles are shown in Tables 4 and 5. These results appear to suggest that CYP2C19*2 and CYP2C19*3 mutations are distinctive in different ethnic groups. Some studies have shown that the CYP2C19*2 and CYP2C19*3 mutant alleles are collaborated with reduced enzyme activity. Clinical studies have shown that patients with CYP2C19*2 allele showed lower levels of the metabolite's activities. This causes a decrease in platelet inhibition activity and higher rates of cardiac occurrences.57 According to various studies, the functional loss of CYP2C19 allelic variants is not in favor of cardiovascular systems following clopidogrel therapeutical regiment.58-60
In a separate study, it was shown that CYP2C19 allelic variants functional loss did not have any adverse effect on the safety and efficacy of clopidogrel among patients with acute coronary diseases.61 Comparison of the Turkman ethnic group with other ethnic populations indicates differences and resemblances in the distribution of CYP2C19 allele and PM genotype. The differences may be associated with racial origin, geographical distribution and environmental factors, etc. Differences in the frequency of CYP2C19 polymorphism in different populations have epidemiologic importance. The frequency of CYP2C19*2 is reported to be around 10% worldwide. Studies have shown that the frequency of CYP2C19*2 generally increases sequentially from Western Asia, Iran to India and Melanesian (with higher than 75% rate). On the other hand, CYP2C19*3 frequency exhibits the same direction. The increase begins in Eastern Asia with the highest prevalency in Melanesia with 33%, but it has been obeserved that its frequency in other part of the world is very low (lower and/or equal to 1%). The frequency of CYP2C19 variant can be as high as 90% in other regions of the world.62
CYP2C19 polymorphism is related to metabolism of some important drugs.55,56 CYP 2C19 phenotypes are reported to affect clinical benefit of several drugs, such as proton pump inhibitors, clopitogrel, sertraline, escitalopram, moclobemide and voriconazol.63 Gastric acid-related abnormalities can be treated with the drug inhibitors and the proton pump inhibitors are sequentially metabolized by CYP2C19 in the liver. CYP2C19 from different genetic backgrounds metabolize the above mentioned drug differently and studies have shown that Japanese and Caucasians poor metabolizers respond properly to the drug with therapeutic dosage, but extensive metabolizers have been shown to have no effect on the above drug at therapeutic dosage.64-66 In a recent clinical trial study, the advantage of CYP2C19 genotype sub-population in having a good response to the proton pump inhibitor in therapeutic dosage was confirmed.67 The finding from another study indicated that Asian populations represent slower metabolism of diazepam than Caucasians. This may depend on the high frequency of the mutant CYP2C19*2 and CYP2C19*3 alleles in Asian populations.56 Toxic doses of diazepam may arise as the result of slower metabolism in PMs. Thus, extra care must be taken with the dosage of diazepam in Asian populations. Subjects with the variants CYP2C19*2 or CYP2C19*3 can denomstrate abnormality in metabolism for drugs such as diazepam, with adverse drug reactions.
The present study showed that PM genotype frequency was highly (10.7%) expressed among the Turkman ethnic group when compared with West Asian,32-34 Europeans,33,35-39 North and South Americans,32,40,41 as well as Scandinavians,42-44 and Africans.45-49 While PM genotype frequency in the Turkman ethnic group was lower than some East Asian27-32 and Oceanian51,52 populations. Moreover, our study showed that the frequency of poor metabolizers in the Turkman ethnic group (10.7%) is relatively close to Thai (9.2%) and Burmese (11%) populations.30
Conclusion
This study confirms the ethnic differences in the CYP2C19 allele and genotype frequencies. Our results also showed that the determined allelic variants of CYP2C19 (CYP2C19*2 and CYP2C19*3 mutations) in the Turkman ethnic group are comparable to other populations. Overall, the determination of CYP2C19 variants in different ethnic groups can be very useful for clinicians to determine the optimal dosage and efficacy of drugs metabolized by this polymorphic enzyme.
Acknowledgements
The authors would like to thank the personnel at the Metabolic Disorders Research Center, the Research Deputy of Golestan University of Medical Sciences for their cooperation and financial support.
References
1. Daly AK. Pharmacogenetics of the major polymorphic metabolizing enzymes. Fundam Clin Pharmacol 2003 Feb;17(1):27-41.
2. Ingelman-Sundberg M. Pharmacogenetics: an opportunity for a safer and more efficient pharmacotherapy. J Intern Med 2001 Sep;250(3):186-200.
3. Daly AK, Cholerton S, Gregory W, Idle JR. Metabolic polymorphisms. Pharmacol Ther 1993 Feb-Mar;57(2-3):129-160.
4. Bertilsson L, Dahl ML, Ingelman-Sundberg M, Johansson I, Sjoqvist F. Inter-individual and inter-ethnic differences in polymorphic drug oxidation. Implication for drug therapy with focus on psychoactive drugs. In: Pacifici GM, Fracchia GN, editors. Advanced in drug metabolism in man. Bruxelles: European Communities; 1995; p. 86–136.
5. Meyer UA. Pharmacogenetics: the slow, the rapid, and the ultrarapid. Proc Natl Acad Sci U S A 1994 Mar;91(6):1983-1984.
6. Meyer UA. In: Carruthers GS, Hoffmann BB, Melmon KL, Nieremberg DW, editors. Drugs in special patient groups: clinical importance of genomics in drug effects. New York: McGraw Hill; 2000.p. 1179–205. Eastern Oriental subjects: Japanese versus mainland Chinese. Clin Pharmacol Ther 1989;46:198–207.
7. Alván G, Bechtel P, Iselius L, Gundert-Remy U. Hydroxylation polymorphisms of debrisoquine and mephenytoin in European populations. Eur J Clin Pharmacol 1990;39(6):533-537.
8. Wilkinson GR, Guengerich FP, Branch RA. Genetic polymorphism of S mephenytoin hydroxylation. In: KalowW, editor. Pharmacogenetics of drug metabolism. New York: Pergamon Press; 1992. p. 657–85.
9. Nakamura K, Goto F, Ray WA, McAllister CB, Jacqz E, Wilkinson GR, et al. Interethnic differences in genetic polymorphism of debrisoquin and mephenytoin hydroxylation between Japanese and Caucasian populations. Clin Pharmacol Ther 1985 Oct;38(4):402-408.
10. Jurima M, Inaba T, Kadar D, Kalow W. Genetic polymorphism of mephenytoin p(4′)-hydroxylation: difference between Orientals and Caucasians. Br J Clin Pharmacol 1985 Apr;19(4):483-487.
11. Bertilsson L, Lou Y-Q, Du Y-L, Liu Y, Kuang T-Y, Liao XM, et al. Pronounced differences between native Chinese and Swedish populations in the polymorphic hydroxylations of debrisoquin and S-mephenytoin. Clin Pharmacol Ther 1992 Apr;51(4):388-397.
12. Horai Y, Nakano M, Ishizaki T, Ishikawa K, Zhou HH, Zhou BI, et al. Metoprolol and mephenytoin oxidation polymorphisms in Far Eastern Oriental subjects: Japanese versus mainland Chinese. Clin Pharmacol Ther 1989 Aug;46(2):198-207.
13. Sohn DR, Kusaka M, Ishizaki T, Shin SG, Jang IJ, Shin JG, et al. Incidence of S-mephenytoin hydroxylation deficiency in a Korean population and the interphenotypic differences in diazepam pharmacokinetics. Clin Pharmacol Ther 1992 Aug;52(2):160-169.
14. Roh H-K, Dahl M-L, Tybring G, Yamada H, Cha Y-N, Bertilsson L. CYP2C19 genotype and phenotype determined by omeprazole in a Korean population. Pharmacogenetics 1996 Dec;6(6):547-551.
15. Goldstein JA. Clinical relevance of genetic polymorphisms in the human CYP2C subfamily. Br J Clin Pharmacol 2001 Oct;52(4):349-355.
16. de Morais SM, Wilkinson GR, Blaisdell J, Nakamura K, Meyer UA, Goldstein JA. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem 1994 Jun;269(22):15419-15422.
17. De Morais SM, Wilkinson GR, Blaisdell J, Meyer UA, Nakamura K, Goldstein JA. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol 1994 Oct;46(4):594-598.
18. Chang M, Dahl M-L, Tybring G, Götharson E, Bertilsson L. Use of omeprazole as a probe drug for CYP2C19 phenotype in Swedish Caucasians: comparison with S-mephenytoin hydroxylation phenotype and CYP2C19 genotype. Pharmacogenetics 1995 Dec;5(6):358-363.
19. Brøsen K, de Morais SM, Meyer UA, Goldstein JA. A multifamily study on the relationship between CYP2C19 genotype and s-mephenytoin oxidation phenotype. Pharmacogenetics 1995 Oct;5(5):312-317.
20. Kaneko A, Kaneko O, Taleo G, Björkman A, Kobayakawa T. High frequencies of CYP2C19 mutations and poor metabolism of proguanil in Vanuatu. Lancet 1997 Mar;349(9056):921-922.
21. Lamba JK, Dhiman RK, Kohli KK. CYP2C19 genetic mutations in North Indians. Clin Pharmacol Ther 2000 Sep;68(3):328-335.
22. Jose R, Chandrasekaran A, Sam SS, Gerard N, Chanolean S, Abraham BK, et al. CYP2C9 and CYP2C19 genetic polymorphisms: frequencies in the south Indian population. Fundam Clin Pharmacol 2005 Feb;19(1):101-105.
23. Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988 Feb;16(3):1215.
24. Goldstein JA, Blaisdell J. Genetic tests which identify the principal defects in CYP2C19 responsible for the polymorphism in mephenytoin metabolism. Methods Enzymol 1996;272:210-218.
25. Zand N, Tajik N, Moghaddam AS, Milanian I. Genetic polymorphisms of cytochrome P450 enzymes 2C9 and 2C19 in a healthy Iranian population. Clin Exp Pharmacol Physiol 2007 Jan-Feb;34(1-2):102-105.
26. Azarpira N, Namazi S, Hendijani F, Banan M, Darai M. Investigation of allele and genotype frequencies of CYP2C9, CYP2C19 and VKORC1 in Iran. Pharmacol Rep 2010 Jul-Aug;62(4):740-746.
27. Yamada S, Onda M, Kato S, Matsuda N, Matsuhisa T, Yamada N, et al. Genetic differences in CYP2C19 single nucleotide polymorphisms among four Asian populations. J Gastroenterol 2001 Oct;36(10):669-672.
28. Takakubo F, Kuwano A, Kondo I. Evidence that poor metabolizers of (S)-mephenytoin could be identified by haplotypes of CYP2C19 in Japanese. Pharmacogenetics 1996 Jun;6(3):265-267.
29. Roh HK, Dahl ML, Tybring G, Yamada H, Cha YN, Bertilsson L. CYP2C19 genotype and phenotype determined by omeprazole in a Korean population. Pharmacogenetics 1996 Dec;6(6):547-551.
30. Tassaneeyakul W, Mahatthanatrakul W, Niwatananun K, Na-Bangchang K, Tawalee A, Krikreangsak N, et al. CYP2C19 genetic polymorphism in Thai, Burmese and Karen populations. Drug Metab Pharmacokinet 2006 Aug;21(4):286-290.
31. Pang YS, Wong LP, Lee TC, Mustafa AM, Mohamed Z, Lang CC. Genetic polymorphism of cytochrome P450 2C19 in healthy Malaysian subjects. Br J Clin Pharmacol 2004 Sep;58(3):332-335.
32. Goldstein JA, Ishizaki T, Chiba K, de Morais SM, Bell D, Krahn PM, et al. Frequencies of the defective CYP2C19 alleles responsible for the mephenytoin poor metabolizer phenotype in various Oriental, Caucasian, Saudi Arabian and American black populations. Pharmacogenetics 1997 Feb;7(1):59-64.
33. Aynacioglu AS, Sachse C, Bozkurt A, Kortunay S, Nacak M, Schröder T, et al. Low frequency of defective alleles of cytochrome P450 enzymes 2C19 and 2D6 in the Turkish population. Clin Pharmacol Ther 1999 Aug;66(2):185-192.
34. Sviri S, Shpizen S, Leitersdorf E, Levy M, Caraco Y. Phenotypic-genotypic analysis of CYP2C19 in the Jewish Israeli population. Clin Pharmacol Ther 1999 Mar;65(3):275-282.
35. Gaikovitch EA, Cascorbi I, Mrozikiewicz PM, Brockmöller J, Frötschl R, Köpke K, et al. Polymorphisms of drug-metabolizing enzymes CYP2C9, CYP2C19, CYP2D6, CYP1A1, NAT2 and of P-glycoprotein in a Russian population. Eur J Clin Pharmacol 2003 Aug;59(4):303-312.
36. Scordo MG, Caputi AP, D’Arrigo C, Fava G, Spina E. Allele and genotype frequencies of CYP2C9, CYP2C19 and CYP2D6 in an Italian population. Pharmacol Res 2004 Aug;50(2):195-200.
37. Bo_ina N, Graniæ P, Laliæ Z, Tramisˇak I, Lovriæ M, Stavljeniæ- Rukavina A. Genetic polymorphisms of cytochromes P450: CYP2C9, CYP2C19, and CYP2D6 in Croatian Population. Roatian Med J 2003;44(4):425-428.
38. Allabi AC, Gala JL, Desager JP, Heusterspreute M, Horsmans Y. Genetic polymorphisms of CYP2C9 and CYP2C19 in the Beninese and Belgian populations. Br J Clin Pharmacol 2003 Dec;56(6):653-657.
39. Ruas JL, Lechner MC. Allele frequency of CYP2C19 in a Portuguese population. Pharmacogenetics 1997 Aug;7(4):333-335.
40. Nowak MP, Sellers EM, Tyndale RF. Canadian Native Indians exhibit unique CYP2A6 and CYP2C19 mutant allele frequencies. Clin Pharmacol Ther 1998 Oct;64(4):378-383.
41. Bravo-Villalta HV, Yamamoto K, Nakamura K, Bayá A, Okada Y, Horiuchi R. Genetic polymorphism of CYP2C9 and CYP2C19 in a Bolivian population: an investigative and comparative study. Eur J Clin Pharmacol 2005 May;61(3):179-184.
42. Halling J, Petersen MS, Damkier P, Nielsen F, Grandjean P, Weihe P, et al. Polymorphism of CYP2D6, CYP2C19, CYP2C9 and CYP2C8 in the Faroese population. Eur J Clin Pharmacol 2005 Aug;61(7):491-497.
43. Yamada H, Dahl M-L, Lannfelt L, Viitanen M, Winblad B, Sjöqvist F. CYP2D6 and CYP2C19 genotypes in an elderly Swedish population. Eur J Clin Pharmacol 1998 Aug;54(6):479-481.
44. Bathum L, Andersen-Ranberg K, Boldsen J, Brøsen K, Jeune B. Genotypes for the cytochrome P450 enzymes CYP2D6 and CYP2C19 in human longevitY. Role of CYP2D6 and CYP2C19 in longevity. Eur J Clin Pharmacol 1998 Jul;54(5):427-430.
45. Xie HG, Kim RB, Stein CM, Wilkinson GR, Wood AJ. Genetic polymorphism of (S)-mephenytoin 4′-hydroxylation in populations of African descent. Br J Clin Pharmacol 1999 Sep;48(3):402-408.
46. Hamdy SI, Hiratsuka M, Narahara K, El-Enany M, Moursi N, Ahmed MS, et al. Allele and genotype frequencies of polymorphic cytochromes P450 (CYP2C9, CYP2C19, CYP2E1) and dihydropyrimidine dehydrogenase (DPYD) in the Egyptian population. Br J Clin Pharmacol 2002 Jun;53(6):596-603.
47. Persson I, Aklillu E, Rodrigues F, Bertilsson L, Ingelman-Sundberg M. S-mephenytoin hydroxylation phenotype and CYP2C19 genotype among Ethiopians. Pharmacogenetics 1996 Dec;6(6):521-526.
48. Herrlin K, Massele AY, Jande M, Alm C, Tybring G, Abdi YA, et al. Bantu Tanzanians have a decreased capacity to metabolize omeprazole and mephenytoin in relation to their CYP2C19 genotype. Clin Pharmacol Ther 1998 Oct;64(4):391-401.
49. Masimirembwa C, Bertilsson L, Johansson I, Hasler JA, Ingelman-Sundberg M. Phenotyping and genotyping of S-mephenytoin hydroxylase (cytochrome P450 2C19) in a Shona population of Zimbabwe. Clin Pharmacol Ther 1995 Jun;57(6):656-661.
50. Dandara C, Masimirembwa CM, Magimba A, Sayi J, Kaaya S, Sommers DK, et al. Genetic polymorphism of CYP2D6 and CYP2C19 in east- and southern African populations including psychiatric patients. Eur J Clin Pharmacol 2001 Apr;57(1):11-17.
51. Kaneko A, Lum JK, Yaviong L, Takahashi N, Ishizaki T, Bertilsson L, et al. High and variable frequencies of CYP2C19 mutations: medical consequences of poor drug metabolism in Vanuatu and other Pacific islands. Pharmacogenetics 1999 Oct;9(5):581-590.
52. Griese EU, Ilett KF, Kitteringham NR, Eichelbaum M, Powell H, Spargo RM, et al. Allele and genotype frequencies of polymorphic cytochromes P4502D6, 2C19 and 2E1 in aborigines from western Australia. Pharmacogenetics 2001 Feb;11(1):69-76.
53. Wedlund PJ, Aslanian WS, McAllister CB, Wilkinson GR, Branch RA. Mephenytoin hydroxylation deficiency in Caucasians: frequency of a new oxidative drug metabolism polymorphism. Clin Pharmacol Ther 1984 Dec;36(6):773-780.
54. Goldstein JA, Faletto MB, Romkes-Sparks M, Sullivan T, Kitareewan S, Raucy JL, et al. Evidence that CYP2C19 is the major (S)-mephenytoin 4′-hydroxylase in humans. Biochemistry 1994 Feb;33(7):1743-1752.
55. Wilkinson GR, Guengerich FP, Branch RA. Genetic polymorphism of S-mephenytoin hydroxylation. Pharmacol Ther 1989;43(1):53-76.
56. Bertilsson L. Geographical/interracial differences in polymorphic drug oxidation. Current state of knowledge of cytochromes P450 (CYP) 2D6 and 2C19. Clin Pharmacokinet 1995 Sep;29(3):192-209.
57. Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med 2009 Jan;360(4):354-362.
58. Shuldiner AR, O’Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA 2009 Aug;302(8):849-857.
59. Simon T, Verstuyft C, Mary-Krause M, Quteineh L, Drouet E, Méneveau N, et al; French Registry of Acute ST-Elevation and Non-ST-Elevation Myocardial Infarction (FAST-MI) Investigators. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med 2009 Jan;360(4):363-375.
60. Collet JP, Hulot JS, Pena A, Villard E, Esteve JB, Silvain J, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction: a cohort study. Lancet 2009 Jan;373(9660):309-317.
61. Paré G, Mehta SR, Yusuf S, Anand SS, Connolly SJ, Hirsh J, et al. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med 2010 Oct;363(18):1704-1714.
62. Sistonen J, Fuselli S, Palo JU, Chauhan N, Padh H, Sajantila A. Pharmacogenetic variation at CYP2C9, CYP2C19, and CYP2D6 at global and microgeographic scales. Pharmacogenet Genomics 2009 Feb;19(2):170-179.
63. Swen JJ, Nijenhuis M, de Boer A, Grandia L, Maitland-van der Zee AH, Mulder H, et al. Pharmacogenetics: from bench to byte–an update of guidelines. Clin Pharmacol Ther 2011 May;89(5):662-673.
64. Furuta T, Ohashi K, Kosuge K, Zhao XJ, Takashima M, Kimura M, et al. CYP2C19 genotype status and effect of omeprazole on intragastric pH in humans. Clin Pharmacol Ther 1999 May;65(5):552-561.
65. Furuta T, Sugimoto M, Shirai N, Ishizaki T. CYP2C19 pharmacogenomics associated with therapy of Helicobacter pylori infection and gastro-esophageal reflux diseases with a proton pump inhibitor. Pharmacogenomics 2007 Sep;8(9):1199-1210.
66. Hunfeld NG, Mathot RA, Touw DJ, van Schaik RH, Mulder PG, Franck PF, et al. Effect of CYP2C19*2 and *17 mutations on pharmacodynamics and kinetics of proton pump inhibitors in Caucasians. Br J Clin Pharmacol 2008 May;65(5):752-760.
67. Furuta T, Shirai N, Kodaira M, Sugimoto M, Nogaki A, Kuriyama S, et al. Pharmacogenomics-based tailored versus standard therapeutic regimen for eradication of H. pylori. Clin Pharmacol Ther 2007 Apr;81(4):521-528.
|
|