The Staphylococcus genus belongs to the Micrococcaceae family. These bacteria are characteristically Gram-positive, non-motile, non-spore forming, and aerobic or facultatively anaerobic.1 More than 20 species have been recognized in this genus, which are distributed in various habitats. Some colonize the skin surface, cutaneous glands, and human or animal (such as ruminants) mucosal membranes. In animals, such as birds and mammals, the skin and the mucous membranes (especially those of the nasopharyngeal region) are considered as the primary reservoirs for Staphylococcus aureus (S. aureus); thereby, this microorganism has enough potential to exist and transmit to or contaminate and spoil animal products such as milk, dairy products, and meats. This is especially true of handled foods during the processing, preparing, wrapping, mincing, and storage steps, which require manipulation. Generally, due to the fact that Staphylococcus can grow in any condition without special or complicated requirements, a high variety of foodstuffs can be contaminated by this microorganism, including meat and meat products especially hamburgers, milk and dairy, chicken, fish, salted and fermented products, and even cooked foods (which account for 24% of all cases). In addition, this microorganism can transmit to environmental sources such as soil, sand, dust, and surface waters.2-4
S. aureus is capable of producing one or more of a family of enterotoxins that known as Staphylococcal enterotoxins (SEs). SEs are classified to classic enterotoxins (SEA–SEE) and new enterotoxins (SEG–SEQ and SER–SEU) based on their biological and serological characteristics. From these twenty types of SEs, A to E are highly isolated from outbreaks of food poisoning in more than 90% of cases. According to studies performed in USA and UK, SEA and SEB have been isolated in 69% of food poisoning cases, while new enterotoxins were causes in 5% of cases.5,6 SEs are considered significant virulence factors belonging to a large family of pyrogenic toxin super antigens (PTSAgs).7
In order to detect SEs from isolated strains and food, several phenotypic methods have been introduced including immunological methods such as agglutination and biological and chromatographic assays. However, the major limitations of these methods are their low accuracy and lack of comprehensiveness. For example, in agglutination due to the fact that different SEs serotypes are partly similar antigenically, cross reactive reactions can occur. In addition, there are only five serologic kits (for SEA to SEE serotypes) available commercially. Therefore, using molecular assays such as the polymerase chain reaction (PCR) can be more accurate and useful than phenotypic methods for detecting enterotoxin producing genes in S. aureus strains.8,9
Methicillin-resistant S. aureus (MRSA) is considered one of the most common pathogens causing nosocomial infection.10,11 MRSA strains are categorized into two families. The first family is known as the community-acquired MRSA (CA-MRSA) strains and are associated with strains acquired in community settings and are expected to be sensitive for many antibiotics. The second family are the hospital-acquired MRSA strains which originate from hospital infections and are resistant to many antibiotics. Foodstuffs have been considered as sources of MRSA strains, therefore, surveying and surveillance activities for foodstuffs harboring MRSA strains is needed.12-14
We aim to evaluate the prevalence of S. aureus strains in food samples of animal origin and to examine their antibacterial susceptibility pattern as well as to use PCR for the molecular detection of the SEs genes and the mecA gene in the isolated strains.
Methods
A total of 1050 food samples were purchased and collected between September 2013 and June 2014, in Hamadan, Iran. The samples were kept in an insulated ice box and transferred to the laboratory and examined within 1–2 hours for the presence of S. aureus according to the Iran national standard (number=1194) for S. aureus identification.
Processing of collected samples for isolation of S. aureus was carried out as follows: approximately 10g of each meat samples were added to test tubes containing 90ml sterile buffered peptone water (BPW; Merck Co., Darmstadt, Germany) and homogenized using vortex mixer (other samples used specific protocols). Then 1ml of each yielded suspension inoculated in test tube containing enrichment media of Giolitti Cantoni broth (QUELAB Co., UK) supplemented with 3.5% potassium tellurite solution and incubated aerobically at 37°C for 24 and 48 hours. The test tubes presenting blackening throughout or in the bottom of the tube were subcultured to Baird-Parker Agar medium (BPA; Merck Co., Darmstadt, Germany) supplemented with 5% egg yolk tellurite emulsion (Merck Co., Darmstadt, Germany) and incubated aerobically at 37°C for 48 hours. Colonies with typical black appearance surrounded by a clearing zone were considered as Staphylococcus genus and were subcultured to blood agar (Merck, Co., Darmstadt, Germany). Colonies that grew in blood agar were identified as S. aureus with some phenotypic criteria. Finally, all presumed identified staphylococcal strains were tested for coagulating citrated rabbit plasma. S. aureus ATCC 25423 was used as positive control and Staphylococcus epidermidis as a negative control.4
Total DNA was extracted from overnight-grown pure isolates using the DNA extraction Kit (BioFlux Co., Tokyo, Japan), according to the manufacturer’s instructions. The quality of extracted DNA was assessed spectrophotometrically by the Nanodrop ND-1000 (Nanodrop Technologies, Inc., Delaware, USA) using 2% agarose gel in 1x TBE buffer.
Following the DNA extraction, to confirm all presumed identified isolates as S. aureus species, a PCR assay targeting the S. aureus-specific nuc gene coding deoxyribonuclease enzyme1 was performed. The reaction mixture for PCR assay was 20μL and was prepared as follows: 10μl of 2x Taq premix Mastermix (Parstous Biotech Co., Iran), 5μl of sterile double-distilled water, 1μl of forward primer, 1μl of reverse primer, and 3μl of DNA sample. The DNA samples as well as a positive control (S. aureus ATCC 25423) and a negative control (S. epidermidis) were amplified by an initial denaturation step for 5 minutes at 94°C followed by 35 cycles of 94°C for 60 seconds, 50°C for 60 seconds, 72°C for 1 minute, and a final extension step at 72°C for 10 minutes in a Bio-Rad Thermal Cycler (Bio-Rad Laboratories, Inc., USA).15 After the amplification reaction, 5μl of each PCR product were visualized and photographed with an UV transilluminator (VilbertLourmat Co., Japan)
For all confirmed S. aureus isolates, antibacterial susceptibility testing were carried out by standard Kirby-Bauer disk agar diffusion (DAD) on Mueller Hinton agar (Merck Co., Germany) according to the Clinical and Laboratory Standards Institute (CLSI, 2013)14 for the antibiotics including cefoxitin (30μg), gentamicin (30μg), erythromycin (15μg), ciprofloxacin (5μg), rifampin (5μg), trimethoprim-sulfamethoxazole, trimethoprim-sulfamethoxazole (SXT) (1.25/23.75μg), clindamycin (2μg), and tetracycline (30μg). All antibiotics were from MAST Co., England. Methicillin-resistance was examined using cefoxitin DAD and confirmed by mecA-specific PCR assay.14
The PCR assay targeting mecA gene coding methicillin-resistance was performed for all isolates.15,16 The reaction mixture for the single target amplification PCR assay was performed as previously described.
Table 1: Primer sequences used for the polymerase chain reaction (PCR) assay in this study.
|
nuc |
F: GCGATTGATGGTGATACGGTT
R: AGCCAAGCCTTGACGAACTAAAGC |
DQ399678 |
279 |
50 |
Hennekinne et al,1 |
|
mecA |
F: GTAGAAATGACTGAACGTCCGATAA
R: CCAATTCCACATTGTTTCGGTCTAA |
X52593 |
310 |
55 |
Hennekinne et al,1 |
|
Multiplex I |
|
sea |
F: GAAAAAAGTCTGAATTGCAGGGAACA
R: CAAATAAATCGTAATTAACCGAAGGTTC |
M18970 |
560 |
55 |
Argudín MÁet al,3 |
|
sei |
F: GTACCGTTGAAAATTCAG
R: AGGCAGTCCATCTCCTG |
AAW3643/
AF285760 |
461 |
55 |
Bianchi et al,4 |
|
seg |
F:TCTCCACCTGTTGAAGG
R: AAGTGATTGTCTATTGTCG |
AF285760 |
323 |
55 |
Bianchi et al,4 |
|
Multiplex II |
|
see |
F: CAAAGAAATGCTTTAAGCAATCTTAGGC
R: CACCTTACCGCCAAAGCTG |
M21319 |
482 |
55 |
Argudín et al,3 |
|
sec |
F:CTTGTATGTATGGAGGAATAACAAAACATG
R: CATATCATACCAAAAAGTATTGCCGT |
X05815 |
275 |
55 |
Argudín et al,3 |
|
seh |
F: CAATCACATCATATGCGAAAGCAG
R: CATCTACCCAAACATTAGCACC |
U11702 |
376 |
55 |
Argudín et al,3 |
|
Multiplex III |
|
sed |
F:GAATTAAGTAGTACCGCGCTAAATAATATG
R: GCTGTATTTTTCCTCCGAGAGT |
M28521 |
492 |
55 |
Argudín et al,3 |
|
seb |
F:ATTCTATTAAGGACACTAAGTTAGGGA
R: ATCCCGTTTCATAAGGCGAGT |
M11118 |
404 |
55 |
Argudín et al,3 |
Three multiplex PCR assays targeting a total of nine SEs genes were designed: multiplex I containing sea, sei, and seg; multiplex II containing see, sec, and seh; and multiplex III containing seb, sed, and sej primer pairs. All primer sequences have been listed in Table 1. Each multiplex PCR reaction carried out in a volume of 25μl that contained: 12.5μl of 2x Taq premix Mastermix, 3.5μl sterile double distilled water, 1μl of each forward primers, 1μl of each reverse primers, and 3μl of DNA sample. In each PCR reaction, the DNA samples as well as a negative control were amplified by an initial denaturation of DNA at 95 °C for 5 minutes and was followed by 35 cycles of amplification (95°C for 30s, 55°C for 30s and 72°C for 60s), and a final extension at 72°C for 10 minutes.17 The amplified PCR products were resolved using electrophoresis and visualized as previously above described.17
For confirmation of PCR product, one sample of each of the enterotoxins gene and mecA PCR products (amplicons) were sequenced (Bioneer Co., Korea) and the data were analyzed using the Chromas software.
All categorical (continuous) variables were compared using the 2-tailed chi-square test or Fisher’s exact test; p-values of ≤0.050 were considered statistically significant. All statistical analyses were performed using the SPSS (SPPS Inc., Chicago, USA) version 20 software package.
Table 2: Information of the collected samples and their polymerase chain reaction (PCR) assay results.
| |
|
|
|
sea |
seb |
sec |
sed |
see |
seg |
seh |
sei |
sej |
sec + sed |
sea + seg |
sed + see |
sea + see + seg |
sea + sed + see + seg |
Total samples with se (%) |
|
Cream |
66 |
4
(6.1) |
0 |
1 |
0 |
0 |
1 |
0 |
1 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
4
(100.0) |
|
Traditional yoghurt |
45 |
2
(4.4) |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1
(50.0) |
|
Raw milk |
271 |
29
(10.7) |
3 |
5 |
1 |
2 |
2 |
1 |
4 |
0 |
2 |
0 |
0 |
2 |
0 |
3 |
0 |
22
(75.9) |
|
Raw red meat |
243 |
25
(10.3) |
0 |
3 |
0 |
1 |
2 |
3 |
6 |
0 |
0 |
0 |
0 |
1 |
1 |
1 |
1 |
19
(76.0) |
|
Raw poultry |
136 |
11
(8.1) |
0 |
2 |
0 |
0 |
1 |
1 |
3 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
8
(72.7) |
|
Traditional cream |
47 |
2
(4.3) |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
1
(50.0) |
|
Cheese |
170 |
19
(11.2) |
2 |
0 |
1 |
1 |
2 |
3 |
3 |
1 |
1 |
0 |
0 |
1 |
0 |
3 |
0 |
16
(84.2) |
|
Butter |
72 |
6
(8.3) |
0 |
0 |
2 |
0 |
0 |
0 |
2 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
5
(83.3) |
Results
A total of 98 (9.3%) S. aureus strains from 1050 food samples were isolated, identified and confirmed by PCR assay targeting the nuc gene [Figure 1]. They included 36 (9.5%) isolates from the 379 raw meat samples and 62 (9.2%) isolates from the 671 raw milk and dairy product samples. The total number of S. aureus isolated among the various collected food type samples has been shown in Table 2.
Table 3 shows the antimicrobial susceptibility patterns of the 98 S. aureus isolates examined. The mecA gene was detected in all MRSA strains, which composed 5.1% of all S.aureus strains [Figure 1].
Prevalence of SEs was determined to be 77.6% (n=76); 79.0% (n=49) in raw milk and dairy products and 75.0% (n=27) in raw meat samples. The percentages of enterotoxigenic strains isolated can be found in Table 2.
The genes of classical enterotoxins (sea, seb, sec, sed, and see) were carried in 63 (64.3%) isolates and those of new enterotoxins (seg, seh, sei, and sej) were carried in 39 (39.8%) of isolates [Figure 1]. Among the genes that code classic enterotoxins, sea gene was the most frequent and was carried by 25 (25.5%) isolates [Table 2].
All our sequenced strains with related PCR product showed the same DNA sequences therefore all PCR assay results were confirmed.
Table 3: Results of antimicrobial susceptibility of Staphylococcus aureus using disk diffusion method.
|
Cefoxitin |
5 (5.1) |
nd |
93 (94.9) |
|
Erythromycin |
30 (30.6) |
3 (3.1) |
65 (66.3) |
|
Tetracycline |
29 (29.6) |
1 (1.0) |
68 (69.4) |
|
Gentamicin |
27 (27.6) |
nd |
71 (72.4) |
|
Clindamycin |
26 (26.5) |
1 (1.0) |
71 (72.4) |
|
Ciprofloxacin |
24 (24.5) |
nd |
74 (75.5) |
|
Rifampin |
24 (24.5) |
4 (4.1) |
70 (71.4) |
nd: not detected; SXT: trimethoprim sulfamethazole
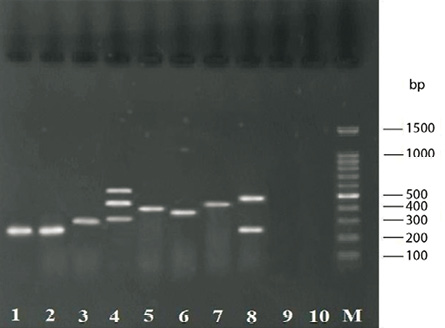
Figure 1: Gel electrophoresis for PCR assays products. Lane M: 100-bp DNA size marker; lane 1: 279-bp amplicon of nuc for S. aureus positive control (ATCC 25423); lane 2: 279-bp amplicon of nuc for food sample; lane 3: 310-bp amplicon for mecA; lane 4: 323-, 482- and 560-bp amplicons for seg, see and sea, respectively; lane 5: 404-bp amplicon for seb; lane 6: 376-bp amplicon for seh; lane 7: 461-bp amplicon for sei; lane 8: 275- and 492-bp amplicons for sec and sed, respectively; line 9: S. epidermidis (negative control); and lane 10: sample without DNA template (negative control).
Discussion
A wide variety of enterotoxin and enterotoxin-like coding genes (more than 20 SEs) have been recognized to have a significant role in stages of host colonizing, gastroenteritis infections, and invasion of skin, mucus, and host defence mechanisms.3,17,18
Food is regarded a significant factor for transferring and spreading of antibiotic resistance genes. S. aureus carrying virulence and antibiotic resistance genes on mobile genetic elements such as plasmids, prophages, and staphylococcal pathogenic islands (SaPIs), and these can horizontally transfer between strains. Consequently, investigation of the antibiotic resistance patterns of S. aureus strains is essential in planning MRSA control measures.19,20
In our study, we aimed to determine the prevalence of enterotoxin producing S. aureus strains collected from foods of animal origin and characterize them. Several phenotypic and genotypic methods were used to detect the presence of enterotoxin and antibiotic resistance genes and to determine the antibacterial susceptibility pattern.
The 9.3% prevalence rate of S. aureus in our study was similar to a study performed in Tehran, which reported a prevalence of 9.5%.21 A study performed in Turkey reported the prevalence of S. aureus in food samples was 13.8%.22 In another study in Canada between 2007 and 2010, the prevalence of S. aureus was 10.5%.23
The contamination rate of raw meat and dairy product samples (9.5% and 9.2%, respectively) was not statistically significant (p>0.050). However, a study by Dallal et al,21 reported a significant difference with a prevalence of 17.1% for dairy products and 3.5% for meat and other products. In a study performed in Italy by Bianchi et al,4 the prevalence of S. aureus was reported as 39% in milk and dairy products,which was higher than our study. A study in Turkey reported a prevalence of 57.3%.24
This inconsistency in the reported prevalence of S. aureus between meat and dairy product samples can be explained by contaminated milk and dairy products via cows and/or sheep with mastitis and during the pasteurization step by human/environmental sources or instruments and equipment with a low standard of cleanliness. Furthermore, cheeses produced from raw and unpasteurized milk are considered significant agents in the spread of S. aureus food poisoning.
Of the 98 S. aureus strains examined in this study, five (5.1%) harbored the mecA gene. Each mecA positive strain demonstrated resistance to cefoxitin (i.e. CA-MRSA). All CA-MRSA strains were isolated from dairy product samples rather than raw meat samples (p<0.050). The following rates of CA-MRSA were reported in other similar studies; 6.4% in Canada, 3.8% in Italy, 2.5% in the Netherlands, 1.6% in Spain, and 2.4% in Korea.23
According to several studies, the main factor in selecting for antibiotic-resistant bacteria is prescribing antibiotics in animal husbandry for therapeutic purposes or as growth promoters. Multiple antimicrobial resistance patterns have been found in S. aureus.14,23
There was no statistically significant difference between raw meat and dairy product samples in antibacterial susceptibility patterns. In addition, statistical analysis comparing antibiotic resistance patterns between CA-MRSA and methicillin-sensitive S. aureus (MSSA) isolates revealed that the CA-MRSA strains were significantly more resistant to antibiotics erythromycin, tetracycline, gentamicin and rifampin than the MSSA strains. This finding was in agreement with a study performed by Dejing et al,25 in 2010. Although resistance to antibiotics ciprofloxacin and SXT were higher in the CA-MRSA strains than that of MSSA strains these differences were not statistically significant. Clindamycin was the only exception as the resistance rate in MSSA strains were in some cases higher than that of CA-MRSA strains.
We assessed gene coding for enterotoxins A–E and G–J amongst the isolated S. aureus. Strains revealed a wide variability in carrying se genes; 77.6% of the strains were carrying one or more of the toxin genes tested. This result was partly consistent with finding in another study that reported 84.9% of foodborne strains harbored enterotoxin.26 In a Portuguese study, the prevalence of enterotoxigenic strains was reported 68.2%9 and another study performed in Italy reported a 59.8% prevalence of se genes in food samples.27 In Japan, Katsuda et al,28 found 183 (67.8%) out of 270 enterotoxigenic S. aureus isolates.
Based on studies, se genes can be located on plasmids (sed and sej), phages (sea and see), pathogenicity islands (seb and sec) and chromosomes (seg, seh, and sei); thereby, several se genes can be harbored by enterotoxigenic S. aureus strains.19
In our study, SEA was the most frequently observed classic SE in enterotoxigenic S. aureus strains. In addition, seg gene was predominant enterotoxin in case of newly identified enterotoxin genes and it was also the most prevalent enterotoxin in our study generally.
SEA, a major leading cause of food poisoning,29,30 is carried by prophage Sa3mu alone or by Sa3mw with other genes (seg and sek).31 In our study, the sea gene was detected in 25.5% (n=25) of S.aureus strains, and also in coexistence with seg gene in 14.3% (n=14). Enterotoxin SEH encoded on the chromosome.19 In our study, it was detected only in one strain isolated from cheese. This finding was consistent with the study performed by Alibayov et al.19 The seg and sei genes are usually detected together19; however, in this study these two genes were detected separately.
In current work, the rate of detected classical enterotoxin genes was higher than that of new enterotoxins (64.3% and 39.8%, respectively). In addition, seven (7.1%), eight (8.2%) and one (1.0%) isolates were simultaneously positive for two, three, and four se genes, respectively. These results are in accordance with findings of the study by Wu et al.25
The detection of SEs genes indicates diversity of the prevalence of se genes in S. aureus strains isolated from animal food samples in Hamadan. In fact, there were differences in the patterns of se genes in relation to the origin of the isolates. It is known that the patterns of SE genes are variable between different geographical origins and years. This difference can be explained by host adaptations of S. aureus in different animal species.13,27,32
There was a significant correlation between S. aureus isolates from butter samples and harboring se genes (p<0.035). However, no significant differences were observed between S. aureus isolates from other food samples and harboring se genes. Additionally, there was no statistically significant difference between raw meat and dairy product samples for harboring se genes.
A statistically significant correlation was found between some antibiotic susceptibility patterns and carrying se genes in examined isolates. This included between cefoxitin and carrying sea and see genes (p=0.001) and between erythromycin and carrying sea, sed, and see genes, tetracycline and carrying sea and see genes, gentamicin and carrying sea gene, rifampin and carrying sec and see genes, and between SXT and carrying sea gene (p<0.050).
Conclusion
S. aureus accounts for frequent contamination of food products and is a well-known clinical and epidemiological pathogen. The detection of the high rate prevalence of enterotoxin genes in this study indicates a potential risk for causing animal originated food poisoning. The increasingly prevalence of CA-MRSA and its emerging antibiotic resistance in foods is a serious problem for public health. Infected animals and infection during the processing stage are the most frequent transmission routes in food contamination with S. aureus or MRSA. Therefore, continuous surveillance and monitoring of pathogens with potential to infect foods and the emergence of MRSA and antibiotic resistance is necessary for public health. For this purpose, the utilization of antibiotics as growth promoters in animal husbandry should be avoided especially those frequently used for caring for both humans and animals.
Disclosure
The authors declared no conflicts of interest.
Acknowledgements
The authors would like to acknowledge the Vice Chancellor of Hamadan University of Medical Sciences for the funding and support of the study.
references
- Hennekinne J-A, De Buyser ML, Dragacci S. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol Rev 2012 Jul;36(4):815-836.
- Akineden O, Hassan AA, Schneider E, Usleber E. Enterotoxigenic properties of Staphylococcus aureus isolated from goats’ milk cheese. Int J Food Microbiol 2008 May;124(2):211-216.
- Argudín MÁ, Mendoza MC, Rodicio MR. Food poisoning and Staphylococcus aureus enterotoxins. Toxins (Basel) 2010 Jul;2(7):1751-1773.
- Bianchi DM, Gallina S, Bellio A, Chiesa F, Civera T, Decastelli L. Enterotoxin gene profiles of Staphylococcus aureus isolated from milk and dairy products in Italy. Lett Appl Microbiol 2014 Feb;58(2):190-196.
- Tang J-n, Tang C, Wang Y, Chen J, Liu J, Liu L, et al. Surveillance study of enterotoxin genes in Staphylococcus aureus isolates from goats of different slaughterhouses in Sichuan, China. Ann Microbiol 2012;62(3):1247-1253.
- Fusco V, Quero GM, Morea M, Blaiotta G, Visconti A. Rapid and reliable identification of Staphylococcus aureus harbouring the enterotoxin gene cluster (egc) and quantitative detection in raw milk by real time PCR. Int J Food Microbiol 2011 Jan;144(3):528-537.
- Jeyasekaran G, Raj KT, Shakila RJ, Thangarani AJ, Karthika S, Luzi M. Simultaneous detection of Staphylococcus aureus enterotoxin C-producing strains from clinical and environmental samples by multiplex PCR assay. Ann Microbiol 2011 Sep;61(3):585-590.
- Ahmady M, Kazemi S. Detection of the enterotoxigenic genes (sei,sej) in Staphylococcus aureus isolates from bovine mastitis milk in the West Azerbaijan of Iran. Comp Clin Path 2013 Jul;22(4):649-654.
- Pereira V, Lopes C, Castro A, Silva J, Gibbs P, Teixeira P. Characterization for enterotoxin production, virulence factors, and antibiotic susceptibility of Staphylococcus aureus isolates from various foods in Portugal. Food Microbiol 2009 May;26(3):278-282.
- David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 2010 Jul;23(3):616-687.
- Shore AC, Coleman DC. Staphylococcal cassette chromosome mec: recent advances and new insights. Int J Med Microbiol 2013 Aug;303(6-7):350-359.
- Yamamoto T, Hung W-C, Takano T, Nishiyama A. Genetic nature and virulence of community-associated methicillin-resistant Staphylococcus aureus. Biomedicine 2013 Jan;3(1):2-18.
- David MZ, Taylor A, Lynfield R, Boxrud DJ, Short G, Zychowski D, et al. Comparing pulsed-field gel electrophoresis with multilocus sequence typing, spa typing, staphylococcal cassette chromosome mec (SCCmec) typing, and PCR for panton-valentine leukocidin, arcA, and opp3 in methicillin-resistant Staphylococcus aureus isolates at a U.S. Medical Center. J Clin Microbiol 2013 Mar;51(3):814-819.
- Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. CLSI document M100-S23. Wayne, PA. Clinical and Laboratory Standards Institute. 2013 Jan;33(1):29-37.
- Zhang K, Sparling J, Chow BL, Elsayed S, Hussain Z, Church DL, et al. New quadriplex PCR assay for detection of methicillin and mupirocin resistance and simultaneous discrimination of Staphylococcus aureus from coagulase-negative staphylococci. J Clin Microbiol 2004 Nov;42(11):4947-4955.
- Zhang K, McClure J-A, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol 2005 Oct;43(10):5026-5033.
- Wu D, Li X, Yang Y, Zheng Y, Wang C, Deng L, et al. Superantigen gene profiles and presence of exfoliative toxin genes in community-acquired meticillin-resistant Staphylococcus aureus isolated from Chinese children. J Med Microbiol 2011 Jan;60(Pt 1):35-45.
- Hwang SY, Kim SH, Jang EJ, Kwon NH, Park YK, Koo HC, et al. Novel multiplex PCR for the detection of the Staphylococcus aureus superantigen and its application to raw meat isolates in Korea. Int J Food Microbiol 2007 Jun;117(1):99-105.
- Alibayov B, Zdeňková K, Purkrtová S, Demnerová K, Karpíšková R. Detection of some phenotypic and genotypic characteristics of Staphylococcus aureus isolated from food items in the Czech Republic. Ann Microbiol 2014;64(4):1587-1596.
- Malachowa N, DeLeo FR. Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci 2010 Sep;67(18):3057-3071.
- Dallal MM, Salehipour Z, Eshraghi S, Mehrabadi JF, Bakhtiari R. Occurrence and molecular characterization of Staphylococcus aureus strains isolated from meat and dairy products by PCR-RFLP. Ann Microbiol 2010;60(2):189-196.
- Aydin A, Sudagidan M, Muratoglu K. Prevalence of staphylococcal enterotoxins, toxin genes and genetic-relatedness of foodborne Staphylococcus aureus strains isolated in the Marmara Region of Turkey. Int J Food Microbiol 2011 Aug;148(2):99-106.
- Crago B, Ferrato C, Drews SJ, Svenson LW, Tyrrell G, Louie M. Prevalence of Staphylococcus aureus and methicillin-resistant S. aureus (MRSA) in food samples associated with foodborne illness in Alberta, Canada from 2007 to 2010. Food Microbiol 2012 Oct;32(1):202-205.
- Ertas N, Gonulalan Z, Yildirim Y, Kum E. Detection of Staphylococcus aureus enterotoxins in sheep cheese and dairy desserts by multiplex PCR technique. Int J Food Microbiol 2010 Aug;142(1-2):74-77.
- Wu D, Wang Q, Yang Y, Geng W, Wang Q, Yu S, et al. Epidemiology and molecular characteristics of community-associated methicillin-resistant and methicillin-susceptible Staphylococcus aureus from skin/soft tissue infections in a children’s hospital in Beijing, China. Diagn Microbiol Infect Dis 2010 May;67(1):1-8.
- Pu S, Wang F, Ge B. Characterization of toxin genes and antimicrobial susceptibility of Staphylococcus aureus isolates from Louisiana retail meats. Foodborne Pathog Dis 2011 Feb;8(2):299-306.
- Normanno G, La Salandra G, Dambrosio A, Quaglia NC, Corrente M, Parisi A, et al. Occurrence, characterization and antimicrobial resistance of enterotoxigenic Staphylococcus aureus isolated from meat and dairy products. Int J Food Microbiol 2007 Apr;115(3):290-296.
- Katsuda K, Hata E, Kobayashi H, Kohmoto M, Kawashima K, Tsunemitsu H, et al. Molecular typing of Staphylococcus aureus isolated from bovine mastitic milk on the basis of toxin genes and coagulase gene polymorphisms. Vet Microbiol 2005 Feb;105(3-4):301-305.
- Le Loir Y, Baron F, Gautier M. Staphylococcus aureus and food poisoning. Genet Mol Res 2003 Mar;2(1):63-76.
- Balaban N, Rasooly A. Staphylococcal enterotoxins. Int J Food Microbiol 2000 Oct;61(1):1-10.
- Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 2001 Apr;357(9264):1225-1240.
- Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, et al. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human dis-ease. Infect Immun 2002 Feb;70(2):631-641.