Methicillin-resistant Staphylococcus aureus (MRSA) is an important nosocomial pathogen infecting defenseless individuals in hospitals throughout the world.1–3 There are growing numbers of studies implying the etiologic role of coagulase-negative Staphylococci (CoNS) in disease in immunocompromised patients, and the increased prevalence of multidrug-resistant strains.4–6 These species have the ability to survive in medical facilities for months.7 The emergence and rise of antibiotic resistance among Staphylococci is a burden in health care facilities and communities.8
Biocides (antiseptics, disinfectants, and preservatives) are chemical compounds applied to inactivate or destroy microorganisms in various settings (e.g., the health care sector, agriculture and the food industry).9–11 By the mid-1900s, many biocidal compounds were in common use as industrial preservatives and in the medical field.10,11 Today, biocides have become an integral part of the industrialized world and are invaluable compounds in the control of human and animal pathogens.12 Large amounts of biocides are therefore consumed within the different settings, including the medical environment where they are used for disinfection, antisepsis, and cleaning.9
A wide variety of biocidal agents, including quaternary ammonium compounds (QACs), such as benzalkonium chloride (BAC) and benzethonium chloride (BZT) and divalent cations like chlorhexidine digluconate (CHDG) are commonly used in hospitals and healthcare
facilities to decontaminate surfaces, disinfect the hands of hospital personnel, and treat hospital-acquired infections.13,14
The worldwide increase in antimicrobial resistance in bacterial pathogens leading to increased mortality and morbidity in humans highlights the importance of infection control practices and the key role of biocides in healthcare. However, the widespread use of biocides in hospitals has led to concerns about the emergence of disinfectant-resistant bacteria. Excessive use may also result in the appearance of cross-resistance between widely used biocides and antibiotics.7,15 Epidemiological data on antiseptic susceptibility and the distribution of resistance genes are useful for nosocomial infection control. Multidrug efflux systems are possibly the most described resistance mechanism with regard to acquired resistance to biocides.
Antiseptic resistance genes, qacA/B, qacC/D, qacE to qacJ (chlorhexidine-resistant genes) and norA (fluoroquinolone efflux transporter protein) have been identified in Staphylococcus species.2,9,16,17 The qacA/B, smr (Staphylococcal multidrug resistance, also known as qacC/D), and norA genes are found mainly in S. aureus clinical isolates and could be responsible for reduced susceptibility to certain antiseptic agents.2,7–9,18,19 The qacA/B and smr genes are mostly found on plasmids, while norA is located on the S. aureus chromosome.9,19 The qacA/B, detected in both S. aureus and CoNS isolates, confers reduced susceptibility to a wide range of antimicrobial organic cations, including QACs and biguanides. The smr gene encodes a protein that belongs to a small multidrug resistance family and confers reduced susceptibility to QACs and ethidium bromide.2,9,14 Additionally, the chromosomal norA gene confers low-level resistance to hydrophilic fluoroquinolones, such as norfloxacin and levofloxacin, as well as antiseptic agents including QACs.19,20 Other plasmid-borne qac genes, qacG, qacH, and qacJ, have been identified in S. aureus and CoNS isolates and their prevalence rates remain low in human carriage isolates.13,17
The incidence rate of MRSA in our hospital has increased to more than 80% of clinical isolates and the prevalence of methicillin -resistant coagulase-negative staphylococci also rising.21 This study was designed to evaluate the efficacy of three different antiseptic agents, BAC, BZT and, CHDG, which are currently used against S. aureus and CoNS clinical isolates. We also sought to determine the prevalence of the antiseptic resistance genes qac, smr, and norA among these bacteria.
Methods
Valiasr Hospital is a 320-bed university-affiliated therapy center located in Arak, Iran. In a 12-month period, from April 2013 to March 2014, various clinical specimens were collected from admitted patients and transported to the laboratory by brain heart infusion (BHI) broth and cultured. Institutional ethical approval was obtained before study commencement.
The isolates were identified using the API-Staph system (API System; bioMérieux, Paris, France). Standard reference species (S. aureus ATCC 25923, S. epidermidis ATCC 12228, and S. saprophyticus ATCC 15305) were used for quality control. All S. aureus isolates were also assessed for the presence of species-specific 442 bp genomic DNA fragment.22,23
MRSA strains were identified by disk diffusion testing on Muller-Hinton agar plate with a cefoxitin disk (30 µg) and an oxacillin disk (10 µg) (Mast, Merseyside, UK) according to Clinical & Laboratory Standards Institute (CLSI) recommendations.22 The presence of the mecA gene was demonstrated using the polymerase chain reaction (PCR) only for phenotypically oxacillin-resistant isolates.3,23 The reference strains S. aureus ATCC 43300 and S. aureus ATCC 25923 were used as positive and negative controls, respectively.
The minimum inhibitory concentration (MIC) of three BAC, BZT, and CHDG biocides (Sigma-Aldrich, St Louis, USA) were determined in duplicate using the microbroth dilution method in accordance with CLSI guidelines.24 Briefly, MICs were determined by twofold serial dilution of each biocide solution (ranging from 0.25 to 128 µg/mL) in Mueller-Hinton broth (MHB). Each dilution was inoculated with 1/10 dilution of 5 × 106 CFU/mL from fresh culture in MHB set up in a microdilution plate. The plate was then incubated at 37 oC for 24 hours. MICs were determined by observing the presence or absence of growth in each dilution. Isolates were considered susceptible to BAC and BZT with a MIC ≤ 3 mg/L and CHDG with an MIC ≤ 1.0 mg/L and to have reduced susceptibility with an MIC between 1.5 and 3.0 mg/L.14
All Staphylococcus isolates were subjected to genomic DNA purification by a DNA extraction kit (Bioflux, Bioneer, Republic of Korea), as described by the manufacturer. PCR was carried out for detection of qacA/B, qacC/D (smr), qacG, qacH, qacJ, and norA genes using the primer sequences listed in Table 1. The volume of each reaction in the PCR was 25 µl. Each reaction mixture contained 12.5 µl of 2× MasterMix (SinaClon, Iran), including 1× PCR buffer, 1.5 mmol/L MgCl2, 0.15 mmol/L dNTP, and 1.25 IU Taq DNA polymerase, 1 µl of each primer (1 µM), 1 µl of template DNA (0.5 µg), and 9.5 µl of sterile distilled water. PCR was performed in the peqSRAR 96X Universal Thermocycler (PEQLAB Ltd, Southampton, UK) for 30 cycles for all studied genes and based on conditions shown in Table 1. PCR products were analyzed by electrophoresis on a 2% (w/v) agarose gel. Three PCR products from each amplified gene were sent for sequencing (Gene-Fanavaran Co., Tehran, Iran) and confirmed through the basic local alignment search tool (BLAST) before the final analysis.
Pearson’s chi-squared test was performed using SPSS Statistics (SPSS Statistics Inc., Chicago, US) version 18.0 to determine association of categorical variables. Statistical significance was set at p-values < 0.050.
Results
In total, 165 Staphylococcus isolates were collected from the clinical specimens, including wounds (n = 39; 23.6%), blood (n=20; 12.1%), sputum (n = 18; 10.9%), urine (n = 7; 4.2%), abscess (n = 20; 12.1%), synovial fluid 18 (10.9%), tracheal aspirates (n = 32; 19.3%), and ascetic fluid (n = 11; 6.6%) of which 60 (36.3%) were MRSA, 54 (32.7%) were methicillin-sensitive S. aureus (MSSA), and 51 (31%) were CoNS strains. All isolates were sa442 positive [Figure 1a].
The 165 Staphylococcus isolates were screened for BAC, BZT and, CHDG resistance. Distribution of MICs of three biocides against all Staphylococci is shown in Table 2. MICs for BAC and BZT ranged from 0.5 to 64 µg/mL, and for CHDG from 0.25 to 8 µg/mL. MRSA isolates showed less susceptibility towards the three biocides tested.
The MIC inhibiting 90% of isolates (MIC90S) tested for BAC, BZT and CHDG in these bacteria were 64 µg/mL, 16 µg/mL, and 2 µg/mL, respectively. CHDG was the most effective biocide with the MIC50 = 1 µg/mL, and MIC90 = 2 µg/mL against MRSA, and MIC50 = 0.5 µg/mL to MIC90 = 1 µg/mL against both MSSA and CoNS.
The mecA gene was detected in all 60 S. aureus isolates, verifying them as MRSA [Figure 1b]. qacA/B was the most common biocide resistance gene among all 165 Staphylococcus isolates (n = 76; 46%) [Table 2]; which comprised of 38 (63.3%) MRSA, 14 (25.9%) MSSA, and 24 (47%) CoNS [Figure 1c]. Eleven (6.7%) and 24 (14.5%) isolates carried smr and norA genes, respectively [Figure 1d and 1e]. In contrast, other resistance genes such as qacG, qacH, and qacJ were absent in all Staphylococci studied.
The qacA/B and smr genes were detected concomitantly in three (1.8%) isolates, of which one (1.7%), one (1.8%) and one (2.0%) were MRSA, MSSA, and CoNS, respectively. Coexistence of three qacA/B, smr, and norA genes was seen among three (5.0%) MRSA, two (3.7%) MSSA, and two (3.9%) CoNS. In addition, 39 (23.6%) strains of the total 165 Staphylococcus isolates were negative for each studied gene [Table 3].
The association of biocide resistance genes and MIC value in bacteria is shown in [Table 4]. Overall, isolates that harbored more resistance genes had reduced susceptibility to biocides.
Table 1: The primer sequences and cycling parameters for detection of genes tested in this study.
|
sa442 |
F: AATCTTTGTCGGTACACGATATTCTTCACG
R:CGTAATGAGATTTCAGTAGATAATAACAACA |
95 oC, 30 s; 55 oC, 30 s; 72 oC, 30 s |
108 |
21 |
|
mecA |
F:TCCAGATTACAACTTCACCAGG
R:CCACTTCATATCTTGTAACG |
94 oC, 30 s; 53 oC, 30 s; 72 oC, 1 min |
162 |
23 |
|
qacA/B |
F: TGGCTTATACCTATTACCTATGG
R: TGTAGAATATGATAATGGATGAC |
94 oC, 1 min; 52 oC, 1 min; 72 oC, 1 min |
169 |
Study own |
|
qacC/D (smr) |
F: GCCATAAGTACTGAAGTTATTGGA
R: GACTACGGTTGTTAAGACTAAACCT |
95 oC, 35 s; 52 oC, 30 s; 72 oC, 1.5 min |
195 |
2 |
|
qacG |
F: CAACAGAAATAATCGGAACT
R: TACATTTAAGAGCACTACA |
95 oC, 1 min; 48 oC, 45 s; 72 oC, 1 min |
670 |
26 |
|
qacH |
F: ATAGTCAGTGAAGTAATAG
R: AGTGTGATGATCCGAATGT |
95 oC, 1 min; 48 oC, 45 s; 72 oC, 1 min |
550 |
26 |
|
qacJ |
F: CTTATATTTAGTAATAGCG
R: GATCCAAAAACGTTAAGA |
95 oC, 1 min; 48 oC, 45 s; 72 oC, 1 min |
667 |
26 |
Table 2: The MIC results of 165 Staphylococci isolates against three antiseptics.
|
BAC |
|
|
|
|
|
|
|
|
|
|
|
|
|
MRSA
(n = 60) |
0 (0) |
0 (0) |
6 (10.0) |
14 (23.3) |
9 (15.) |
14 (23.3) |
5 (8.3) |
5 (8.3) |
7 (11.7) |
0 (0) |
8 |
64 |
|
MSSA
(n = 54) |
0 (0) |
26 (48.1) |
16 (29.6) |
5
(9.2) |
2 (3.7) |
1
(1.8) |
2 (3.7) |
1 (1.8) |
1
(1.8) |
0 (0) |
1 |
4 |
|
CoNS
(n = 51) |
0 (0) |
15 (29.4) |
0 (0) |
8 (15.7) |
11 (11.7) |
4
(7.8) |
6 (11.7) |
5 (9.8) |
2
(3.9) |
0 (0) |
4 |
32 |
|
BZT |
|
|
|
|
|
|
|
|
|
|
|
|
|
MRSA
(n = 60) |
0 (0) |
4
(6.7) |
10 (16.7) |
14 (23.3) |
9 (15.0) |
9 (15.0) |
10 (16.7) |
1 (1.7) |
3 (5) |
0 (0) |
4 |
16 |
|
MSSA
(n = 54) |
0 (0) |
19 (35.2) |
27 (50.0) |
5
(9.2) |
0 (0) |
1
(18) |
2 (3.7) |
0 (0) |
0 (0) |
0 (0) |
1 |
2 |
|
CoNS
(n = 51) |
0 (0) |
15 (29.4) |
0 (0) |
10 (19.6) |
4 (7.8) |
9 (17.6) |
13 (25.5) |
0 (0) |
0 (0) |
0 (0) |
4 |
16 |
|
CHDG |
|
|
|
|
|
|
|
|
|
|
|
|
|
MRSA
(n = 60) |
0 (0) |
24
(40) |
9 (15.0) |
24 (40.0) |
1 (1.7) |
2
(2.3) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
1 |
2 |
|
MSSA
(n = 54) |
10 (18.5) |
34 (63.0) |
7 (13.0) |
1
(1.8) |
2 (3.7) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0 (0) |
0.5 |
1 |
BAC: benzalkonium chloride; BZT: benzethonium chloride; CHDG: chlorhexidine digluconate; MIC: minimum inhibitory concentration; MRSA: methicillin-resistant S. aureus; MSSA: methicillin-susceptible S. aureus; CoNS: coagulase-negative Staphylococci.
Table 3: Distribution of the biocides resistance genes in 165 Staphylococcus isolates.
|
qacA/B |
37 (61.7) |
15 (27.8) |
24 (47.1) |
76 (46.1) |
|
smr |
6 (10.0) |
2 (3.7) |
3 (5.9) |
11 (6.7) |
|
norA |
10 (16.7) |
12 (22.2) |
2 (3.9) |
24 (14.5) |
|
qacA/B + smr |
1 (1.7) |
1 (1.8) |
1 (2.0) |
3 (1.8) |
|
qacA/B + norA |
2 (3.3) |
0 (0.0) |
1 (2.0) |
3 (1.8) |
|
smr + norA |
1 (1.7) |
0 (0.0) |
1 (2.0) |
2 (1.2) |
|
qacA/B + smr + norA |
3 (5.0) |
2 (3.7) |
2 (3.9) |
7 (4.2) |
|
Negative |
0 (0.0) |
22 (40.7) |
17 (33.3) |
39 (23.6) |
MRSA: methicillin-resistant S. aureus; MSSA: methicillin-susceptible S. aureus; CoNS: coagulase-negative Staphylococci.
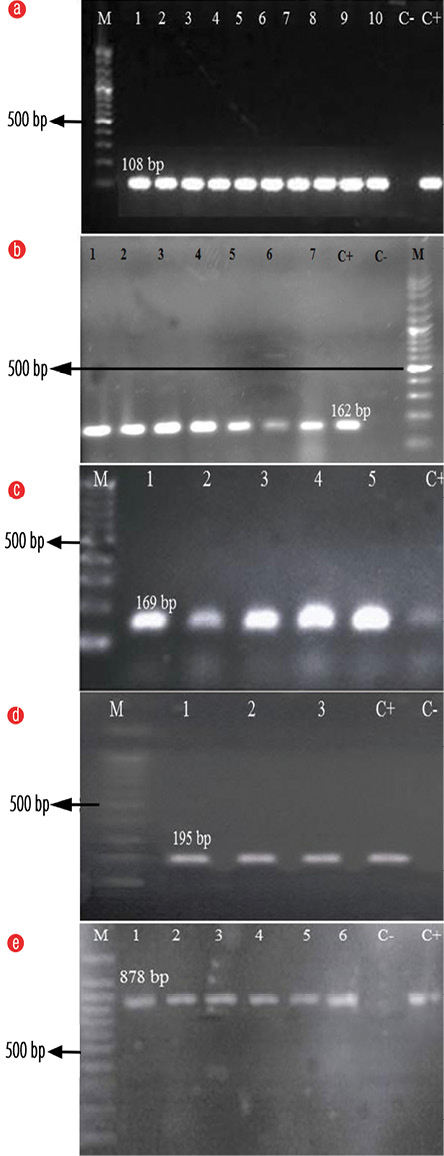
M: 100 bp Plus DNA Ladder; C-: Negative control; C+: Positive control.
Figure 1: Agarose gel electrophoresis of the polymerase chain reaction (PCR) amplification of the (a) sa442, (b) mecA, (c) qacA/B, (d) smr, and (e) norA genes. PCR product of isolates in numbered lanes.
Table 4: Correlation between biocide resistance genes and MIC value in Staphylococcal isolates.
|
qacA/B |
1–64 |
2–32 |
2–32 |
2–16 |
1–16 |
2–16 |
0.5–2 |
0.5–1 |
0.5–1 |
76 |
|
smr |
2–4 |
2–4 |
2 |
1 |
1 |
2 |
0.5 |
1 |
0.5 |
11 |
|
norA |
1–2 |
2 |
0.5 |
0.5–2 |
1–2 |
0.5 |
0.5 |
0.5–1 |
0.5 |
24 |
|
qacA/B+ smr |
4- 32 |
2 |
4 |
2 |
2 |
4 |
2 |
2 |
0.5 |
3 |
|
qacA/B+ norA |
64 |
- |
16 |
64 |
- |
8 |
2 |
- |
1 |
3 |
|
smr+ norA |
4 |
- |
8 |
8 |
- |
4 |
1 |
- |
1 |
2 |
|
qacA/B+smr+norA |
64 |
16–64 |
64 |
32–64 |
8–16 |
4–16 |
4–8 |
4 |
2 |
7 |
|
Negative |
1 |
0.5–1 |
0.5 |
0.5–1 |
0.5–1 |
0.5 |
0.5 |
0.25–0.5 |
0.25–0.5 |
39 |
MIC: minimum inhibitory concentration; BAC: benzalkonium chloride; BZT: benzethonium chloride; CHDG: chlorhexidinedigluconate; MRSA: methicillin-resistant S. aureus; MSSA: methicillin-susceptible S. aureus; CoNS: coagulase-negative staphylococci.
Discussion
The treatment and control of infections associated with Staphylococci, particularly MRSA and also CoNS, has become more difficult due to their ability to acquire resistance to many antimicrobials. This partly resulted from the ability of such bacteria to persist in healthcare environments and transmit among patients. Vigorous infection control measures, including standard hygiene and disinfection with efficient biocides, could lead to a decrease in the circulation of bacterial resistant clones in
hospital environments.
In Iran, there are many reports about the increased antimicrobial resistance in Staphylococcal isolates,3,25,26 but relatively few studies have investigated antiseptic susceptibility. Thus, assessments on the increase or decrease of antiseptic susceptibility in such problematic organisms could provide valuable information to support infection control and prevent healthcare-associated infections.
Our study can be considered the first comprehensive study in Iran evaluating the susceptibility to antiseptic agents and distribution of antiseptic resistance genes of 165 MRSA, MSSA, and CoNS strains collected from a single therapy center in Arak. CLSI guidelines (M100 document, 2015) and related published papers have been used as interpretive criteria for susceptibility testing.5,14 The MICs of three test disinfectants against our Staphylococci isolates were ≤ 64 µg/mL, which is lower than concentrations recommended for use (2000, 1000, and 5000 µg/mL for BAC, BZT, and CHDG, respectively). This would suggest that if these biocides are used in the hospital environment in accordance with the manufacturers’ instructions, 100% of bacteria should be inhibited or killed. However, it is noteworthy that in clinical practice, concentrations that might remain on surfaces after cleaning might provide a selective pressure on microorganisms. In the hospital environment, bacteria grow in biofilms on surfaces that have been shown to provide cells a 10–1000 fold higher tolerance of antimicrobial agents, and may be a contributor to disinfection failure.27
The qacA/B genes alone were found in 46.1% of tested bacteria: 61.7% of MRSA, 27.8% of MSSA, and 47.1% of CoNS. The high prevalence of these genes is likely to be due to selective pressure imposed by the various disinfection agents used in hospitals. Moreover, it is of note that significantly more MRSA than MSSA isolates were positive for the qacA/B gene. This may be the location of the qacA/B genes on widespread multi-resistance plasmids like the pSK1 (qacA) and the β-lactamase/heavy metal resistance families, such as pSK23. These plasmids frequently harbor a number of determinants responsible for resistance to a range of antimicrobial agents.9,28 Since MRSA multi-resistance is also, in part, due to such multi-resistance plasmids, the high incidence of qacA/B in MRSA is apprehensible.
The relatively low rate of carriage of smr (qacC/D) and norA genes found in the present study is in contrast to others. For example, smr was identified in 44.2% and norA in 36.7% of MRSA isolates in a study from the UK.1 This may be due to the larger number of isolates screened and/or variation in the type of biocides used regularly, applying different selective pressures on Staphylococci isolates in each region. In addition, qacG, qacH, and qacJ genes were not detected in our study. These findings support the fact that among Gram-positive bacteria, the qac genes clearly predominate in Staphylococci with qacA/B genes most frequently reported followed by qacC/D genes.15,29 Other qac genes, such as qacG, qacH, and qacJ have been less frequently observed.17 On the other hand, compared to qacA/B, the role of other genes like norA as a contributing factor in the resistance of cationic antiseptic agents is very low.In another study conducted in neonatal intensive care units of a hospital in France from 51 CoNS isolated from catheter-associated bloodstream infections, 21 (41.2%) were resistance to at least one biocide and 30 (59%) isolates were positive for qacA/B resistance genes. This finding also emphasizes the important role of qacA/B genes in biocide resistance.30
We found a significant difference in MIC value between the isolates with and without biocide resistance genes. Furthermore, a considerable association was observed between the coexistence of several genes and the degree of susceptibility to all biocides tested. For example, isolates that carried qacA/B, smr, and norA genes, had MIC values higher than those positive for one or two resistance genes. This means that isolates carrying more biocide resistance genes may be able to persist on human skin and clinical environments where residual concentrations of biocides after cleaning may be lower than in-use concentrations.
Conclusion
At this time, the qacG, qacH, and qacJ resistance genes do not seem to pose a problem among Staphylococcus isolates collected from our hospital. However, the presence of other genes for biocide resistance, especially qacA/B, in the clinical Staphylococcal population and their ability to develop reduced susceptibility highlights the importance of effective infection control strategies, the use of biocides at concentrations recommended by the manufacturer, and the continued surveillance of resistance gene carriage.
Disclosure
The authors declared no conflicts of interest. The study was funded bv the Arak University of Medical Sciences (grant number 92-144-2). This article was extracted from the thesis prepared by Nona Taheri to fulfill the requirements required to earn the Master of Science degree.
references
- 1. Vali L, Davies SE, Lai LL, Dave J, Amyes SG. Frequency of biocide resistance genes, antibiotic resistance and the effect of chlorhexidine exposure on clinical methicillin-resistant Staphylococcus aureus isolates. J Antimicrob Chemother 2008 Mar;61(3):524-532.
- 2. Noguchi N, Nakaminami H, Nishijima S, Kurokawa I, So H, Sasatsu M. Antimicrobial agent of susceptibilities and antiseptic resistance gene distribution among methicillin-resistant Staphylococcus aureus isolates from patients with impetigo and staphylococcal scalded skin syndrome. J Clin Microbiol 2006 Jun;44(6):2119-2125.
- 3. Pournajaf A, Ardebili A, Goudarzi L, Khodabandeh M, Narimani T, Abbaszadeh H. PCR-based identification of methicillin-resistant Staphylococcus aureus strains and their antibiotic resistance profiles. Asian Pac J Trop Biomed 2014 May;4(Suppl 1):S293-S297.
- 4. Barbier F, Ruppé E, Hernandez D, Lebeaux D, Francois P, Felix B, et al. Methicillin-resistant coagulase-negative staphylococci in the community: high homology of SCCmec IVa between Staphylococcus epidermidis and major clones of methicillin-resistant Staphylococcus aureus. J Infect Dis 2010 Jul;202(2):270-281.
- 5. Zhang M, O’Donoghue MM, Ito T, Hiramatsu K, Boost MV. Prevalence of antiseptic-resistance genes in Staphylococcus aureus and coagulase-negative staphylococci colonising nurses and the general population in Hong Kong. J Hosp Infect 2011 Jun;78(2):113-117.
- 6. Sharma V, Jindal N. In vitro activity of vancomycin and teicoplanin against coagulase negative staphylococci. Oman Med J 2011 May;26(3):186-188.
- 7. Zmantar T, Kouidhi B, Miladi H, Bakhrouf A. Detection of macrolide and disinfectant resistance genes in clinical Staphylococcus aureus and coagulase-negative staphylococci. BMC Res Notes 2011;4:453.
- 8. Saadat S, Solhjoo K, Norooz-Nejad MJ, Kazemi A, Van A, Van B. Positive Vancomycin-resistant Staphylococcus aureus Among Clinical Isolates in Shiraz, South of Iran. Oman Med J 2014 Sep;29(5):335-339.
- 9. McDonnell G, Russell AD. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev 1999 Jan;12(1):147-179.
- 10. Wijesinghe LP, Weerasinghe TK. A Study on the Bactericidal Efficiency of Selected Chemical Disinfectants and Antiseptics. OUSL Journal 2010;6:44-58.
- 11. Hugo WB. A brief history of heat and chemical preservation and disinfection. J Appl Bacteriol 1991 Jul;71(1):9-18.
- 12. EU (European Union). Regulation (EU) No 528/2012 of the European Parliament and of the Council of 22 May 2012 concerning the making available on the market and use of biocidal products. Offic J Europ Uni 2012;27:1-123.
- 13. Smith K, Gemmell CG, Hunter IS. The association between biocide tolerance and the presence or absence of qac genes among hospital-acquired and community-acquired MRSA isolates. J Antimicrob Chemother 2008 Jan;61(1):78-84.
- 14. Shamsudin MN, Alreshidi MA, Hamat RA, Alshrari AS, Atshan SS, Neela V. High prevalence of qacA/B carriage among clinical isolates of meticillin-resistant Staphylococcus aureus in Malaysia. J Hosp Infect 2012 Jul;81(3):206-208.
- 15. Longtin J, Seah C, Siebert K, McGeer A, Simor A, Longtin Y, et al. Distribution of antiseptic resistance genes qacA, qacB, and smr in methicillin-resistant Staphylococcus aureus isolated in Toronto, Canada, from 2005 to 2009. Antimicrob Agents Chemother 2011 Jun;55(6):2999-3001.
- 16. Ye JZ, Yu X, Li XS, Sun Y, Li MM, Zhang W, et al. [Antimicrobial resistance characteristics of and disinfectant-resistant gene distribution in Staphylococcus aureus isolates from male urogenital tract infection]. Zhonghua Nan Ke Xue 2014 Jul;20(7):630-636.
- 17. Ye HF, Zhang M, O’Donoghue M, Boost M. Are qacG, qacH and qacJ genes transferring from food isolates to carriage isolates of staphylococci? J Hosp Infect 2012 Jan;80(1):95-96.
- 18. Nakipoğlu Y, Iğnak S, Gürler N, Gürler B. [The prevalence of antiseptic resistance genes (qacA/B and smr) and antibiotic resistance in clinical Staphylococcus aureus strains]. Mikrobiyol Bul 2012 Apr;46(2):180-189.
- 19. Furi L, Ciusa ML, Knight D, Di Lorenzo V, Tocci N, Cirasola D, et al; BIOHYPO Consortium. Evaluation of reduced susceptibility to quaternary ammonium compounds and bisbiguanides in clinical isolates and laboratory-generated mutants of Staphylococcus aureus. Antimicrob Agents Chemother 2013 Aug;57(8):3488-3497.
- 20. Sabatini S, Gosetto F, Iraci N, Barreca ML, Massari S, Sancineto L, et al. Re-evolution of the 2-phenylquinolines: ligand-based design, synthesis, and biological evaluation of a potent new class of Staphylococcus aureus NorA efflux pump inhibitors to combat antimicrobial resistance. J Med Chem 2013 Jun;56(12):4975-4989.
- 21. Rezazadeh M, Yousefi Mashouf R, Sarmadyan H, Ghaznavi-Rad E. Antibiotic profile of methicillin-resistant Staphylococcus aureus with multiple-drug resistances isolated from nosocomial infections in Vali-asr Hospital of Arak. Arak Medic J 2013;16(2):29-37.
- 22. Bode LG, van Wunnik P, Vaessen N, Savelkoul PH, Smeets LC. Rapid detection of methicillin-resistant Staphylococcus aureus in screening samples by relative quantification between the mecA gene and the SA442 gene. J Microbiol Methods 2012 May;89(2):129-132.
- 23. Ghaznavi-Rad E, Shamsudin MN, Sekawi Z, van Belkum A, Neela V. A simplified multiplex PCR assay for fast and easy discrimination of globally distributed staphylococcal cassette chromosome mec types in meticillin-resistant Staphylococcus aureus. J Med Microbiol 2010;59(10): 1135-1139.
- 24. Clinical Laboratory Standards Institute (CLSI). Performance standards for antimicrobial susceptibility testing: twentieth informational supplement. 2015; M100-S25. [cited 2015 Febuary]. Available from http://shop.clsi.org/site/Sample_pdf/M100S25_sample.pdf.
- 25. Pourakbari B, Sadr A, Ashtiani MT, Mamishi S, Dehghani M, Mahmoudi S, et al. Five-year evaluation of the antimicrobial susceptibility patterns of bacteria causing bloodstream infections in Iran. J Infect Dev Ctries 2012 Feb 13;6(2):120-125.
- 26. Askari E, Soleymani F, Arianpoor A, Tabatabai SM, Amini A, Naderinasab M. Epidemiology of mecA-methicillin resistant Staphylococcus aureus (MRSA) in Iran: A systematic review and meta-analysis. Iran J Basic Med Sci 2012 Sep;15(5):1010-1019.
- 27. Gilbert P, Allison DG, McBain AJ. Biofilms in vitro and in vivo: do singular mechanisms imply cross-resistance? J Appl Microbiol 2002;92(Suppl):98S-110S.
- 28. Paulsen IT, Firth N, Skurray RA. Resistance to antimicrobial agents other than b -lactams. W: The Staphylococci in human disease. Red. K. B. Crossley, G. L. Archer. Churchill Livington, New York b 1997;175-212.
- 29. Zmantar T, Kouidhi B, Hentati H, Bakhrouf A. Detection of disinfectant and antibiotic resistance genes in Staphylococcus aureus isolated from the oral cavity of Tunisian children. Ann Microbiol 2012;62(1):123-128.
- 30. Lepainteur M, Royer G, Bourrel AS, Romain O, Duport C, Doucet-Populaire F, et al. Prevalence of resistance to antiseptics and mupirocin among invasive coagulase-negative staphylococci from very preterm neonates in NICU: the creeping threat? J Hosp Infect 2013 Apr;83(4):333-336.